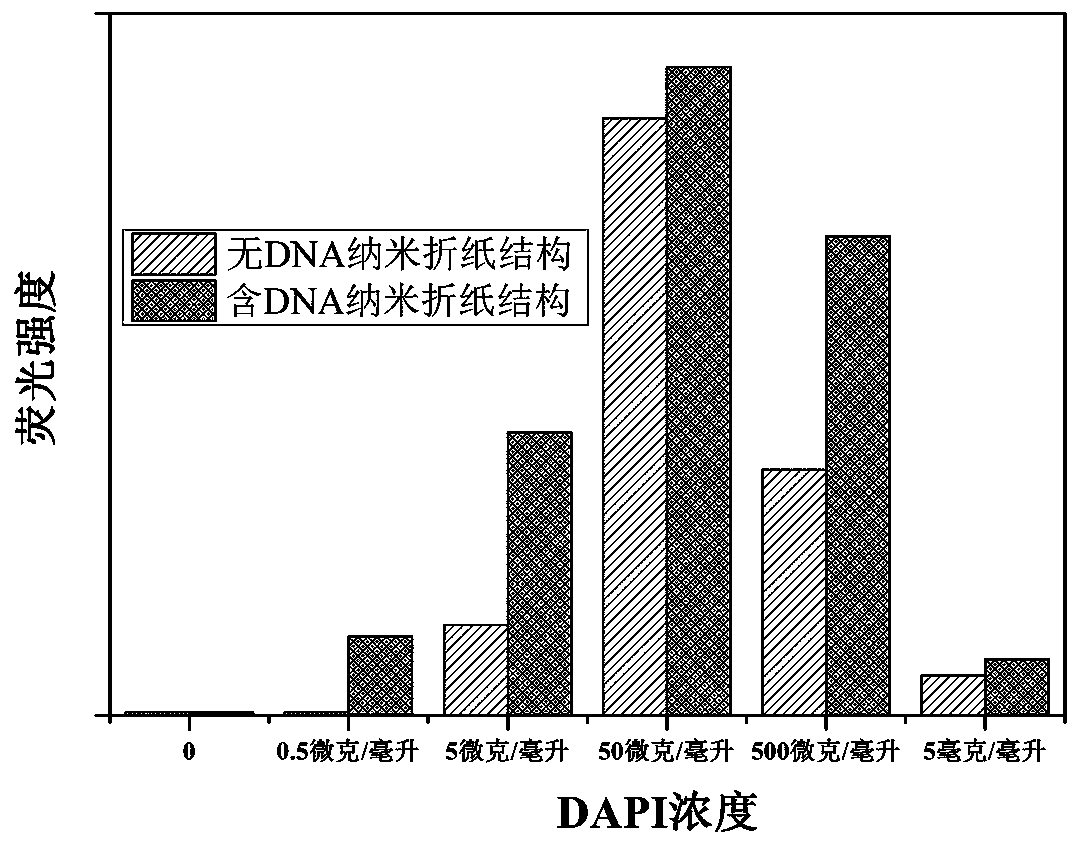
A method for embedding and releasing DNA nano-origami structures as drug carriers using DAPI embedding and releasing
An origami structure and nanotechnology, which can be used in pharmaceutical formulations, medical preparations with inactive ingredients, fluorescence/phosphorescence, etc., and can solve the problems of expensive reagent ordering, complicated experimental design and operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Add 12 μL of circular DNA (2 μM, sequence: 5'TATGCCCAGCCCTGTAAGATGAAGATAGCGCACAATGGTCGGATTCTCAACTCGTATTCTCAACTCGTATTCTCAACTCGTCCTGCCCTμL) to 16 μL of primer DNA (1 μM, sequence: 5'CCAGCCTAAGAGTTGAGCA3'), dNTPs (4 μL, 10 mM), RCA buffer polymerase 4μL, 10 U / μL), the final volume of the mixed solution was 40μL. Then amplified in a 30°C water bath for 30 min. 40 μL of the obtained product was subjected to 10% PAGE electrophoresis, and then recovered by slicing gel to remove small fragments of DNA, redundant circular DNA and primer DNA. Take 3 μL of the recovered product, add 12 μL of milliQ water, and add stapled strands 1-3 (100 μM, the sequence is as follows:
[0016] 5'-CAGCCCTGTAAGATGAAGATAGCGTCTATGCC-3'
[0017] 5'-CCCTGACTCACAATGGTCGGATTCCGTCTCTG-3'
[0018] 5’-TCTCAACTTCAACTCGTATTCTCAACTCGTAT-3’) 1 μL each, 2 μL TAE buffer (10×, 125 mM Mg 2+ ), the total volume is 20 μL, the solution is mixed well, placed in a high temperature of 95 °C, and then gradually cooled...
Embodiment 2
[0020] Add 12 μL of circular DNA (2 μM, sequence: 5'TATGCCCAGCCCTGTAAGATGAAGATAGCGCACAATGGTCGGATTCTCAACTCGTATTCTCAACTCGTATTCTCAACTCGTCCTGCCCTμL) to 16 μL of primer DNA (1 μM, sequence: 5'CCAGCCTAAGAGTTGAGCA3'), dNTPs (4 μL, 10 mM), RCA buffer polymerase 4μL, 10 U / μL), the final volume of the mixed solution was 40μL. Then amplified in a 30°C water bath for 30 min. 40 μL of the obtained product was subjected to 10% PAGE electrophoresis, and then recovered by slicing gel to remove small fragments of DNA, redundant circular DNA and primer DNA. Take 3 μL of the recovered product, add 12 μL of milliQ water, and add stapled strands 1-3 (1 μM, the sequence is as follows:
[0021] 5'CAGCCCTGTAAGATGAAGATAGCGTCTATGCC3'
[0022] 5'CCCTGACTCACAATGGTCGGATTCCGTCTCTG3'
[0023] 5’TCTCAACTTCAACTCGTATTCTCAACTCGTAT3’, 1 μL each, 2 μL TAE buffer (10×, 125mMMg 2+ ), the total volume is 20 μL, the solution is mixed well, placed in a high temperature of 95 °C, and then gradually cooled to 25 °C....
Embodiment 3
[0025]Add 12 μL of circular DNA (2 μM, sequence: 5'TATGCCCAGCCCTGTAAGATGAAGATAGCGCACAATGGTCGGATTCTCAACTCGTATTCTCAACTCGTATTCTCAACTCGTCCTGCCCTμL) to 16 μL of primer DNA (1 μM, sequence: 5'CCAGCCTAAGAGTTGAGCA3'), dNTPs (4 μL, 10 mM), RCA buffer polymerase 4μL, 10 U / μL), the final volume of the mixed solution was 40μL. Then amplified in a 30°C water bath for 30 min. 40 μL of the obtained product was subjected to 10% PAGE electrophoresis, and then recovered by slicing gel to remove small fragments of DNA, redundant circular DNA and primer DNA. Take 3 μL of the recovered product, add 12 μL of milliQ water, and add stapled strands 1-3 (1 μM, the sequence is as follows:
[0026] 5'CAGCCCTGTAAGATGAAGATAGCGTCTATGCC3'
[0027] 5'CCCTGACTCACAATGGTCGGATTCCGTCTCTG3'
[0028] 5’TCTCAACTTCAACTCGTATTCTCAACTCGTAT3’, 1 μL each, 2 μL TAE buffer (10×, 125mMMg 2+ ), the total volume is 20 μL, the solution is mixed well, placed in a high temperature of 95 °C, and then gradually cooled to 25 °C. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com