Nucleic acid membrane strip and reagent kit for non-deletion type alpha-thalassemia gene detection and detection method
A thalassemia and non-deletion technology, applied in the field of non-deletion α-thalassemia gene detection, can solve the problems of increasing the risk of missed detection and non-inclusion, and achieve high specificity, simple operation, and increase amplification efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0066] 1.2 Preparation of nucleic acid membrane strips
[0067] Probe preparation: Dilute the probes with 0.5mol / L carbonate buffer (pH8.4) to the sample concentration, mix well, arrange them in order, and put them in an ice box for later use.
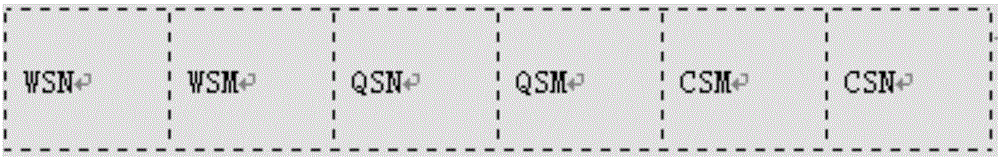
[0068] Membrane strip pre-treatment: Cut the membrane into a size suitable for printing by the printer, and use the printer to print the spotting area as shown in Table 2. Table 2 is the position table of the nucleic acid membrane strip.
[0069] Table 2
[0070] WSN
WSM
QSN
QSM
CSM
CSN
[0071]Soak the membrane with 0.1mol / L HCl solution twice, 5 minutes each time, then soak the membrane with 16% EDAC solution for 15 minutes, and finally wash it with purified water 4 times, 5 minutes each time, and place it at room temperature for more than 30 minutes.
[0072] Spotting the membrane: spot the probes on the corresponding area according to the position of the probes on the membrane, 0.8 μl for each ...
Embodiment 1
[0091] Preparation of sample genomic DNA
[0092] This kit has no specified requirements for the extraction method of human genomic DNA. Traditional methods such as phenol-chloroform method or commercial nucleic acid extraction kits can be used to extract human genomic DNA. It is recommended to use the blood / cell / tissue of Tiangen Biochemical Technology (Beijing) Co., Ltd. Genomic DNA extraction kit, and operate according to the instructions of the kit. Blood samples should be anticoagulated with EDTA or sodium citrate instead of heparin.
[0093] 2. Preparation of nucleic acid membrane strips
[0094] 2.1 Probe preparation
[0095] Dilute the probes with 0.5mol / L carbonate buffer (pH8.4) to the sample concentration, mix well, arrange them in order, and put them in an ice box for later use. The sample concentration of the probe is shown in Table 6. Table 6 is the sample concentration table of probes.
[0096] Table 6
[0097]
[0098] 2.2 Pretreatment of film strips: ...
Embodiment 2
[0117] The hybridization membrane strip and the kit of the present invention detect clinical sample results and compare the gold standard sequencing results.
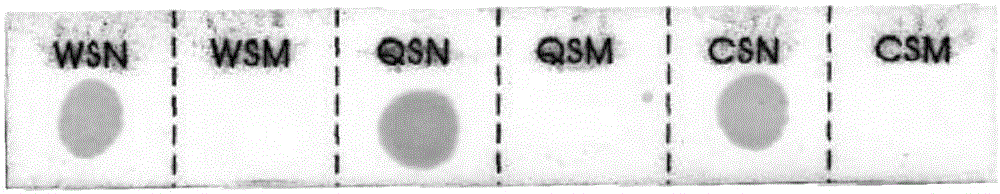
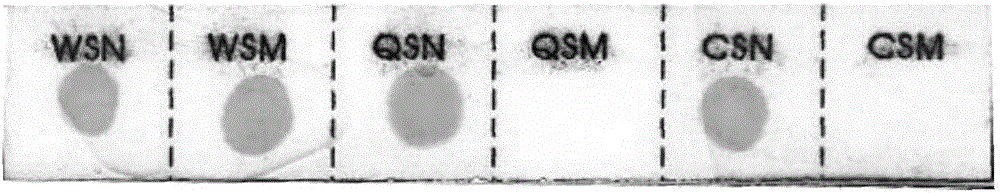
[0118] Select 1248 cases of clinical samples, respectively adopt the hybridization membrane strip and kit of the present invention to detect the mutations of WS, QS, and CS genes in the samples, and detect the α2 genes CD30 (-GAG), CD31 (AGG>AAG), CD43 / 44(-C), CD49(-CG), CD59(GGC>GAC) and WS, QS, CS gene mutations, the detection results are shown in Table 8, with the sequencing results as the standard, the hybridization membrane strip and kit of the present invention detect clinical samples The positive and negative results of the non-deleted α-thalassemia gene are shown in Table 9. Table 8 is a comparison table of the detection results of the hybridization membrane strip and the kit of the present invention and the dideoxy chain termination sequencing method.
[0119] Table 8
[0120]
[0121]
[0122] Table 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com