TaqMan probe primer mixture, kit and fluorescent quantitative PCR detection method for quickly identifying bungarus multicinctus blyth
A technology of the money snake and primer composition, which is applied in the direction of biochemical equipment and methods, microbe determination/testing, DNA/RNA fragments, etc., to achieve good accuracy, reduce the risk of interference, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
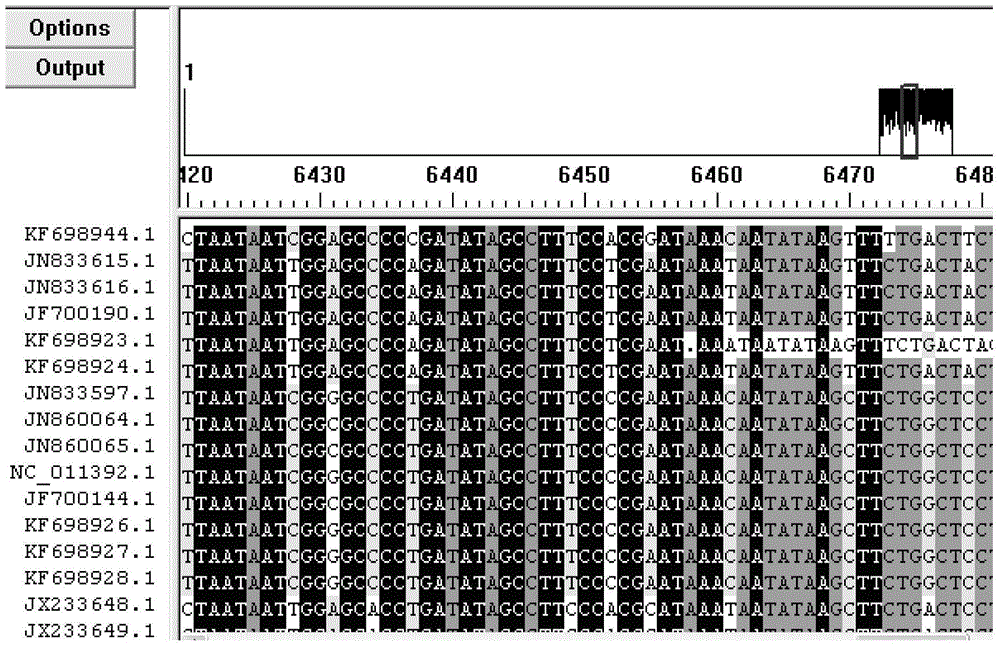
[0052] (1) Primer design: select the cytochrome C gene as the target gene based on DNA barcoding technology (as shown in SEQ ID NO.1), search and download the snake mitochondrial COI gene sequence in Genbank (GeneBank accession numbers are KF698926.1, JF700190.1, KF698937.1, KF698948.1, KF698944.1, JX233650.1, JN833612.1, JX233641.1, JX233637.1), by using the Mega5 analysis software to compare the money snake and common counterfeit red snake, Homology of COI gene sequence of golden krait, cobra, Elaphe japonica, yellow chain snake, lead-colored water snake, Chinese water snake and double whole white krait, design specific primers (as shown in SEQ ID NO.2 and SEQ ID NO.3 Shown) and TaqMan probe (as shown in SEQ ID NO.4), comparison results see figure 1 .
[0053] Design and construct a recombinant plasmid containing positively amplified internal standard DNA, and design the corresponding TaqMan probe (as shown in SEQ ID NO.5), the method is: use DNA random generation software ...
Embodiment 2
[0072] specificity test
[0073] Genomic DNA extracted from the confused Pinchia viper, golden krait, cobra, Elaphe japonica, yellow chain snake, lead-colored water snake, Chinese water snake, Shuangquan white ring snake and authentic money snake as templates, according to Example 1 The optimized reaction system and reaction conditions were used for fluorescent quantitative PCR to detect the specificity of primers and probes.
[0074]The results showed that only the DNA derived from the white snake had a CT value <35, while the other samples had no amplification signal. The results are shown in Table 4. It can be seen that the primers and probes in this kit have strong species specificity.
[0075] Table 4 Fluorescent quantitative PCR detection CT value
[0076]
Embodiment 3
[0077] Embodiment 3 sensitivity test
[0078] Genomic DNA Sensitivity Test
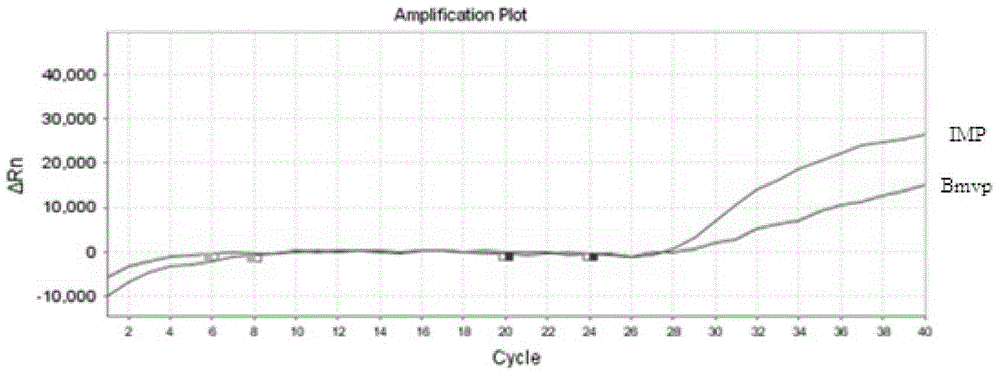
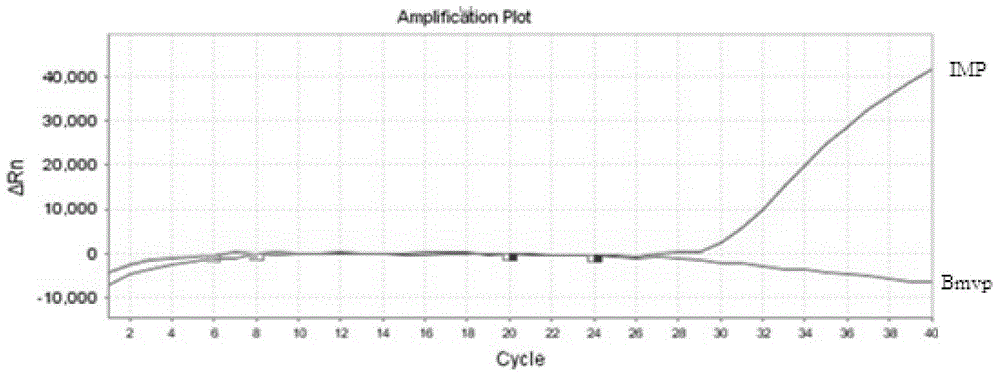
[0079] Extract target genomic DNA according to the kit used in Example 1, quantify to 50ng with Nanodrop, do 10× gradient dilution (10 -1 , 10 -2 , 10 -3 , 10 -4 , 10 -5 ), 2 μl was taken as the template amount for each gradient, and 12 parallel samples were set for each gradient, and the DNA of Snake chrysalis was detected according to the above method to investigate the sensitivity of the kit.
[0080] The results show( Figure 4 , Figure 5 ), when the amount of fluorescent quantitative template DNA is 0.01ng, the probe still has an amplification curve and the CT value is 35, and there is basically no amplification curve, so the kit The detection limit of the method is 0.01ng;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com