A pair of Talen recognition sequences targeting zebrafish forkhead box n1 gene and its mRNA preparation method
A technology for identifying sequences and gene sequences, applied in the field of genetic engineering, can solve the problems that RNAi cannot completely replace, increase the workload of practical operation, and low efficiency of gene homologous recombination, and achieve simple and accurate experimental design, low cost and low toxicity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Preparation of mRNA for targeted knockout of the zebrafish Forkhead box n1 gene
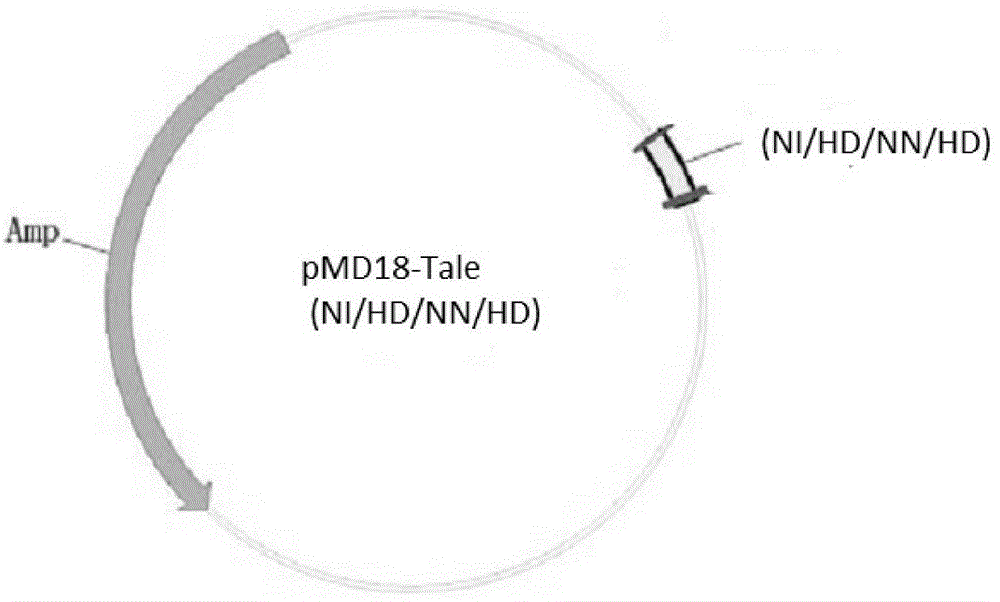
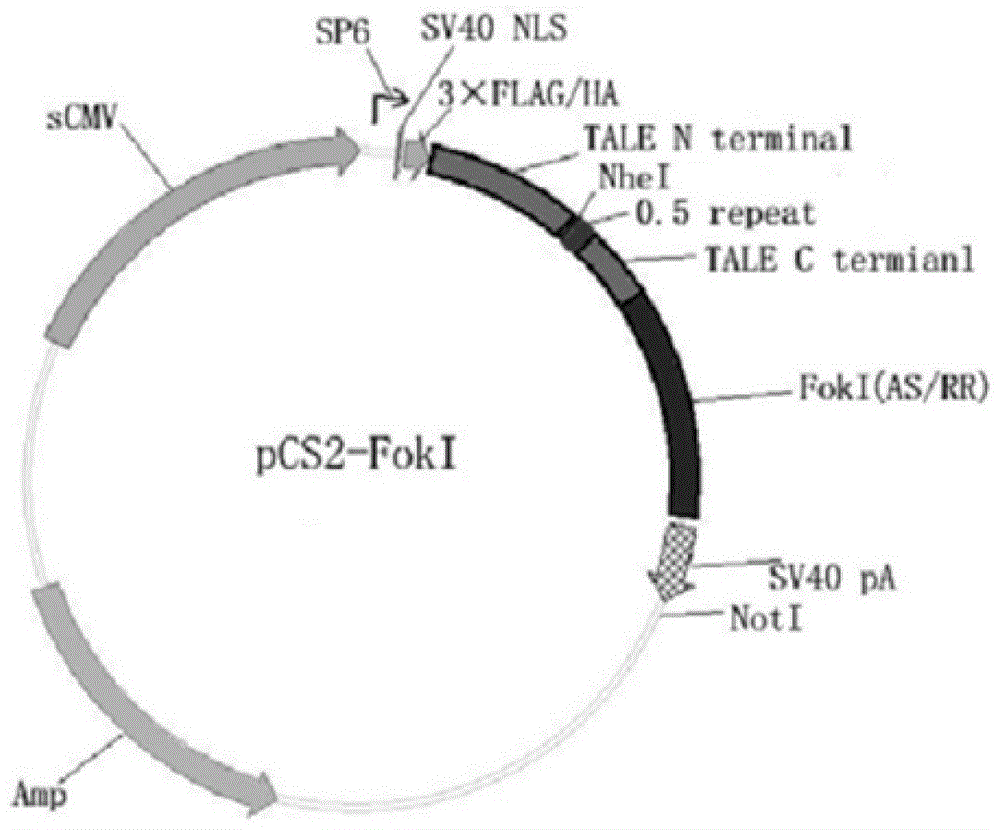
[0038] In the present invention, the materials pMD-18 (NI / HD / NN / HD) and pCS2-Foxn1 are all derived from the construction of Zhang Bo Laboratory of Peking University, 293T cells are derived from the ACTT cell line bank, and the mRNA synthesis kit (mMESSAGE SP6Kit) was purchased from ambion, and the Tu zebrafish were bred in the Zebrafish House, College of Life Sciences, Sun Yat-sen University.
[0039] The experimental methods used in the present invention are conventional experimental methods unless specifically mentioned.
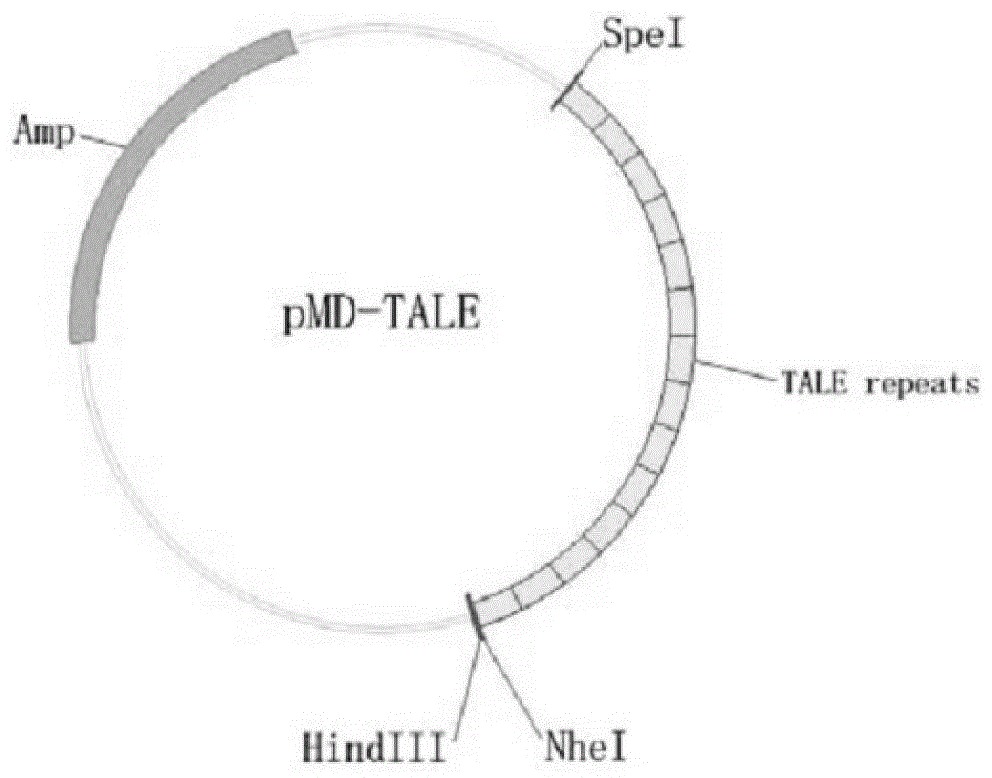
[0040] 1. Construction of talen expression vector targeting Forkhead box n1 gene
[0041] The genome sequence of zebrafish (GenBank: Gene ID: 266748) was detected from the genome database of the NCBI website, and its exon usage was analyzed. Based on the Talen gene knockout principle, the appropriate target sequence Talen recognition target sequence was se...
Embodiment 2
[0059] Example 2: Targeted knockout of Zebrafish Forkhead box n1 gene by Foxn1-Tal-Fok1 mRNA
[0060] 1. Injection of Foxn1-Tal-Fok1 mRNA
[0061] After mixing an appropriate amount of left and right arm mRNA, the combined mRNA is referred to as Foxn1-Tal-Fok1 mRNA, diluted with RNAase-free ddH2O to a final concentration of 100ng / ul, and added 1 / 10 of the volume of phenored to mix well, passed Microinjected into Tu lineage zebrafish embryos.
[0062] 2. Gene variation analysis of embryos injected with Foxn1-Tal-Fok1
[0063] Twenty 3dpf Tu lineage zebrafish embryos injected with foxn1-Tal-Fok1 mRNA were randomly collected and used to extract genomic DNA. The DNA band of the Foxn1 target site was amplified by PCR, and the primers used were:
[0064] TalenFoxn1JD-F0:5'-AAGGCACTATTCAAGGACACCCAGACCCTGG-3',
[0065] TalenFoxn1JD-R1:5'-TCCACATCAGTGCCTAATGTAGTCCAAGAG-3'
[0066] Use Afe1 digestion to analyze the amplification products, such as Figure 6 As shown, the embryonic ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com