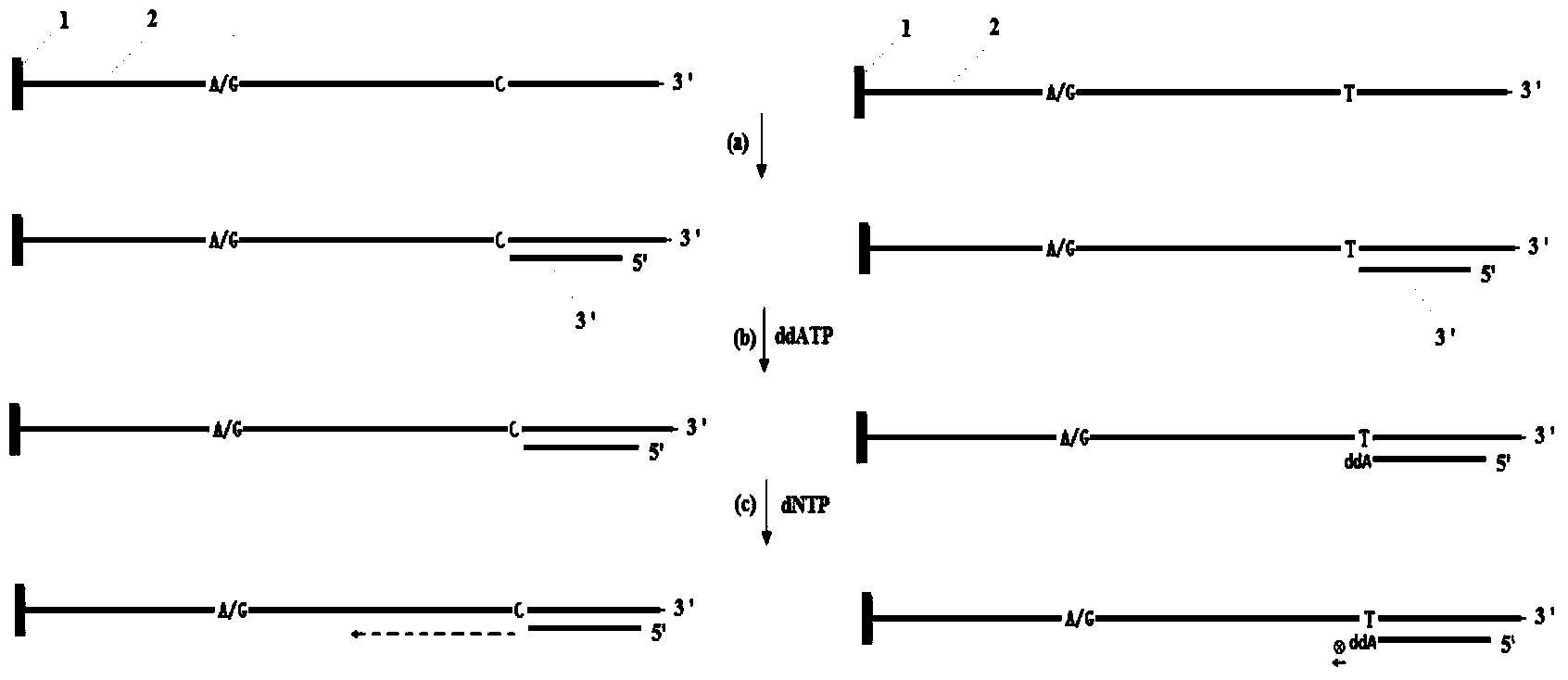Method for analyzing haplotype of PCR products by primer selection sequencing-by-synthesis
A technology of synthetic sequencing and haplotype, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., and can solve the problems of cumbersome operation, different amplification efficiency, and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
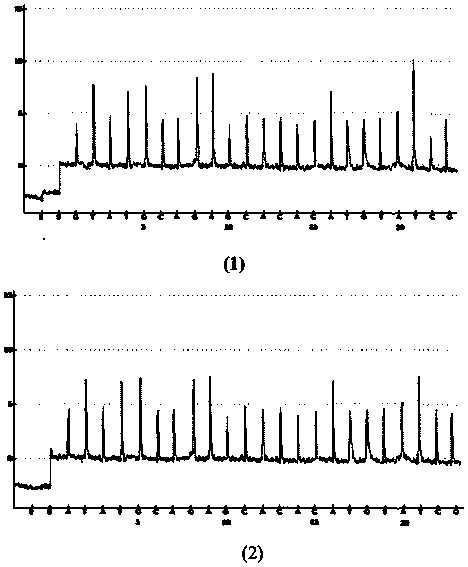
[0034] Example 1: The haplotype analysis of the MMP7 (U25346) gene PCR product comprising A / G (rs11568818) and C / T (rs11568819) nucleic acid fragments
[0035] (1) Genomic DNA in peripheral blood was extracted from the blood sample using traditional protein kinase K and phenol / chloroform extraction methods;
[0036] (2) PCR primers (5'-AGTCTACAGAACTTTGAAAGTATGTG-3', 5'-biotin-CTATGAGAGC AGTCATTTGACTTTG-3', 200 ng genomic DNA, 0.2mM dNTP, 1 U Taq DNA polymerase, 1×amplification buffer, 1.8 mM MgCl 2 50 μL PCR amplification system for amplification. The amplification conditions are: initial denaturation at 94°C for 5 minutes, 35 thermal cycles: denaturation at 94°C for 30 seconds, annealing at 61°C for 45 seconds, extension at 72°C for 45 seconds, and finally extension at 72°C for 7 minutes.
[0037] (3) React the PCR amplification with avidin-modified magnetic beads, immobilize the biotin-modified DNA strand on the magnetic beads, denature it under 0.2M NaOH solution, remove ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com