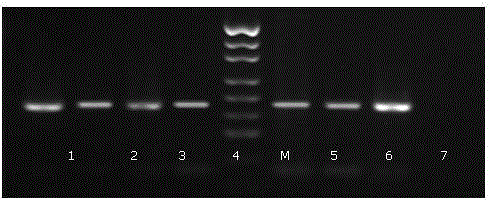
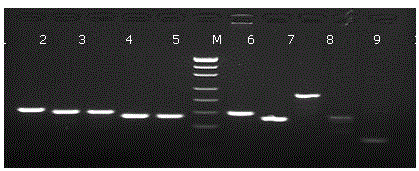
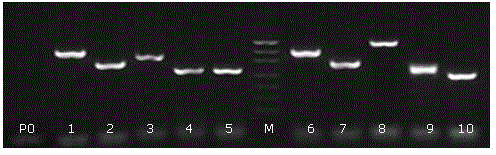
Old animal sample species identification kit and preparation method
An animal sample, obsolete technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as non-human, cumbersome detection process, improper DNA extraction, etc., to promote progress, simple effect, and many solutions. The effect of primer interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Preparation and use of old animal sample species identification kit 1
[0063] 1. Utilize the hypervariable I region sequence of the mammalian mitochondrial genome in Genebank to design two pairs of PCR primers, the sequence of the primers is:
[0064] Primer A: ACGAAACAGGATCCAACAAC
[0065] Primer B: GGTGTAGTTGTCTGGGTCTCC
[0066] Utilize the hypervariable I region sequence of mammalian mitochondrial genome in Genebank to design two pairs of PCR primers, the sequence of described primers is:
[0067] Primer C: CCATCAACACCCAAAGCTG
[0068] Primer D: CATTTAATGCACGACGTACAT
[0069] Primer E: CCCATGCATAATAAGCACGTAC
[0070] Primer F: GAGGCATGGTGATTAAGCTC
[0071] 2. Prepare a kit including the following components: 1 tube of Mastermix reaction solution (1ml / tube), 1 tube of P-mix-1 reaction solution and 1 tube of P-mix-2 reaction solution (0.1ml / tube), DNA molecular weight standard 1 tube (0.5ml / tube).
[0072] 3. Use process:
[0073] (1) Sample DNA:
[0074] Sel...
Embodiment 2
[0087] Preparation and use of old animal sample species identification kit 2
[0088] 1. Utilize the hypervariable I region sequence of the mammalian mitochondrial genome in Genebank to design two pairs of PCR primers, the sequence of the primers is:
[0089] Primer A: ACGAAACAGGATCCAACAAC
[0090] Primer B: GGTGTAGTTGTCTGGGTCTCC
[0091] Utilize the hypervariable I region sequence of mammalian mitochondrial genome in Genebank to design two pairs of PCR primers, the sequence of described primers is:
[0092] Primer C: CCATCAACACCCAAAGCTG
[0093] Primer D: CATTTAATGCACGACGTACAT
[0094] Primer E: CCCATGCATAATAAGCACGTAC
[0095] Primer F: GAGGCATGGTGATTAAGCTC
[0096] 2. Prepare a kit including the following components: 1 tube of Mastermix reaction solution (1ml / tube), 1 tube of P-mix-1 reaction solution and 1 tube of P-mix-2 reaction solution (0.1ml / tube), DNA molecular weight standard 1 tube (0.125ml / tube).
[0097] 3. Specimen collection, preservation and recording:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com