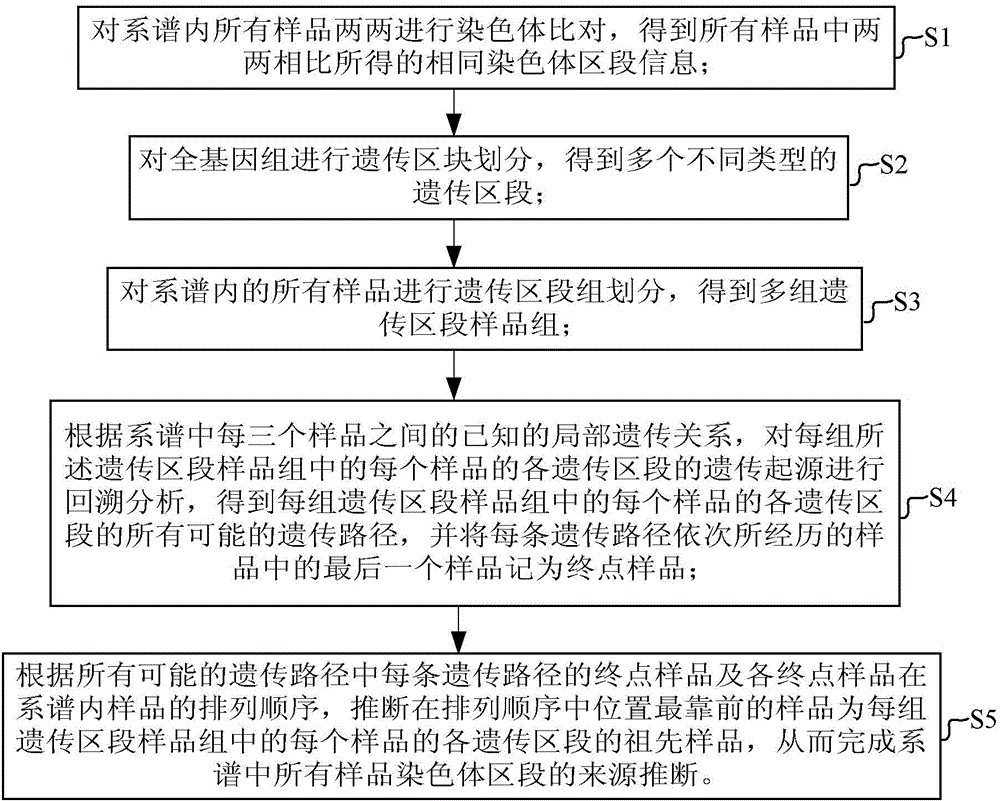
Device and method for inferring sources of chromosome regions in pedigree
A chromosome and pedigree technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of missing sample data information, affecting the coherence and integrity of pedigree, and the inability to know the complete pedigree structure information, etc., to achieve compatibility Good, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
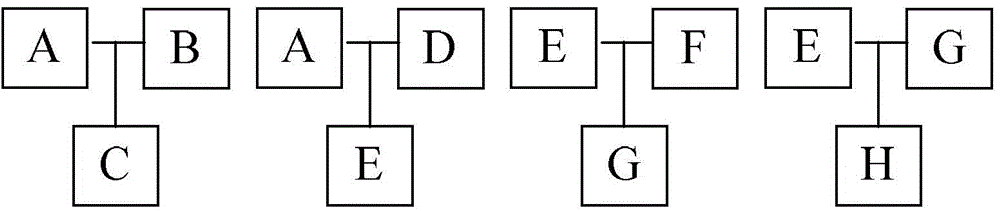
[0044] A rice pedigree, consisting of eight samples of A, B, C, D, E, F, G and H, currently only know the local pedigree of three samples, such as figure 2 As shown, that is, A and B are the parents of C; A and D are the parents of E; E and F are the parents of G; E and G are the parents of H.
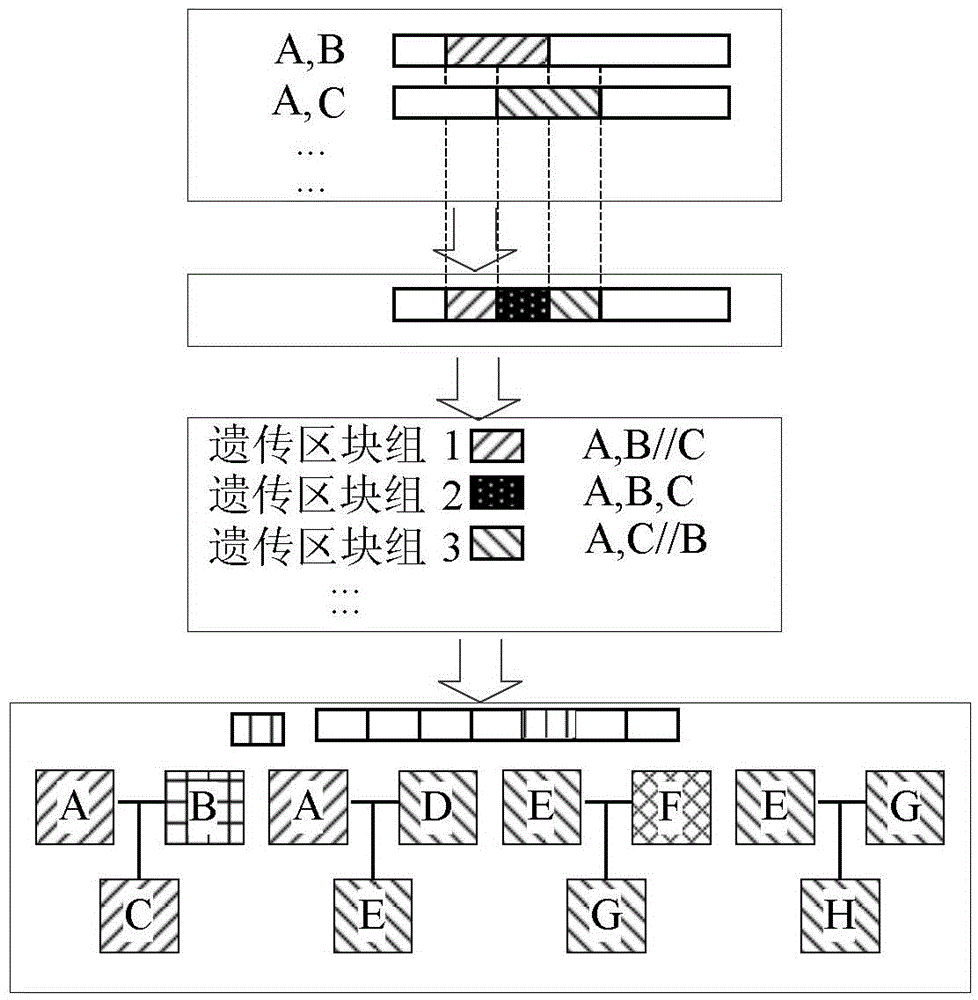
[0045] according to image 3 As shown, by using the IBDseq algorithm to compare the chromosomes of the above 8 samples in pairs, through the comparison of A and B, the same chromosome segment between A and B is found; then A and C are compared to find A and C Between the same chromosome segment, followed by A and D, A and E, and so on, until G and H, to find the same chromosome segment between each two samples in 8 samples; then the whole genome of rice Genetic segments are divided to obtain a plurality of different types of genetic segments; and then according to the types of each genetic segment, the above-mentioned 8 samples are grouped according to the type of each genetic segmen...
Embodiment 2
[0059] The pedigree samples of Example 2 and the steps before obtaining different grouped samples are the same as those in Example 1, and the origin of the fifth genetic segment of each sample is inferred, the difference is that the retrospective analysis method used is different, The backtracking analysis in Example 2 is inferred by recording the path experienced by each sample in the previous step of the search, and the next step of the search path does not need to return to the sample at the starting point, but only needs to return to the bifurcation point and continue to search for the path that has not been experienced. The specific analysis process is as Figure 5 said, where:
[0060] A is the ancestral sample in the pedigree, its theoretical backtracking path is the same as the actual backtracking path, and the type of its genetic segment is still the A sample itself; similarly,
[0061] The genetic segment type of the B sample is also the B sample itself;
[0062] T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com