Biomarkers of response to proteasome inhibitors
A proteasome inhibitor and labeling technology, which is applied in the fields of peptide/protein components, biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problem that patients cannot bear the time of trial and error selection of treatment plans, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
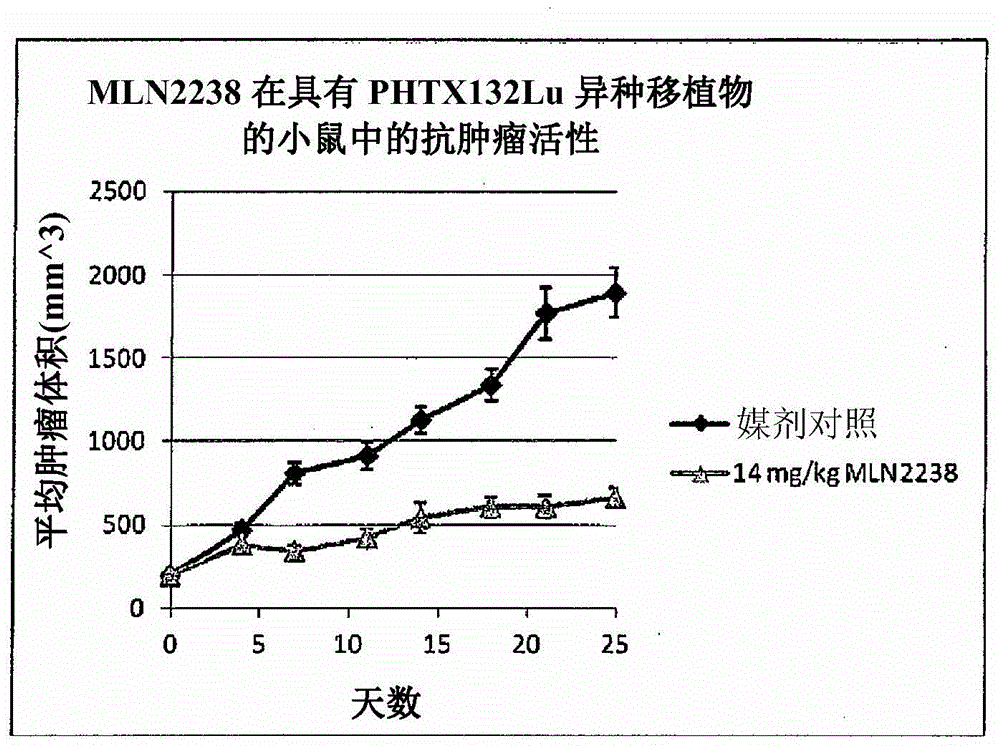
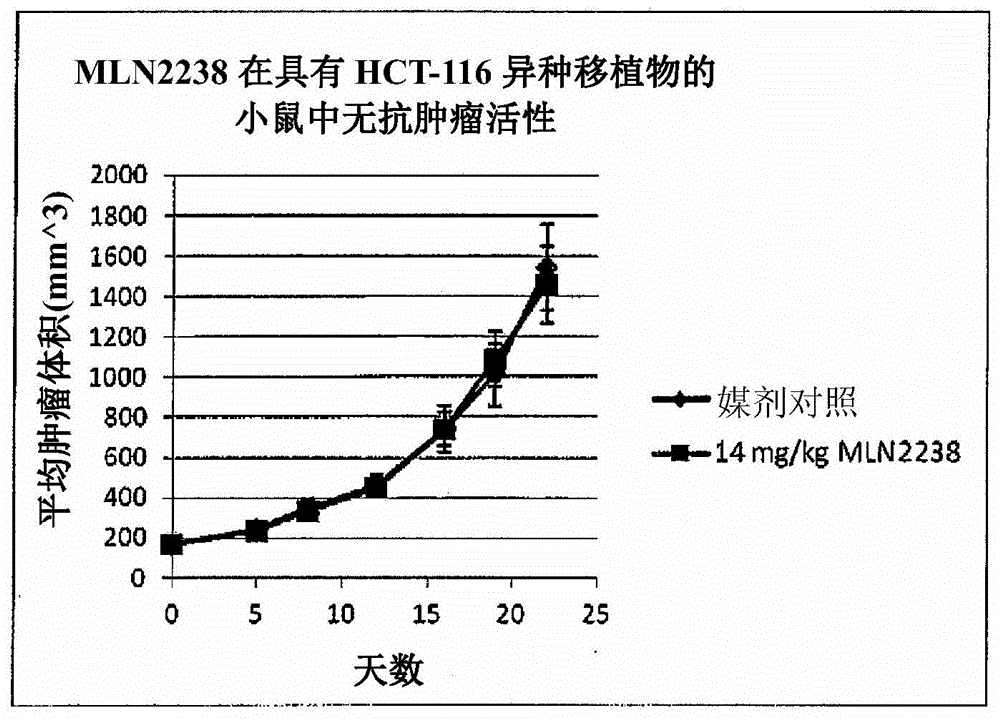
[0226] xenograft
[0227] Xenografts with names beginning with PHTX were obtained at Millennium Pharmaceuticals, Inc. as follows: Tumors obtained from patients were obtained through the Cooperative Human Tissue Network and the National Disease Research Exchange. Research Interchange). Within 24 hours of surgery, tumors were implanted in 4 SCID-NOD mice. Tumors were serially passaged 2-3 times in SCID-NOD mice to confirm growth, and material was stored in liquid nitrogen to retrieve tumors for future use. Tumors were further passaged into a large number of NCr-nude and / or CB17 SCID mice for study as MLN2238 listed in Table 3. Tumors taken from patients were histologically characterized by H&E staining, and DNA was prepared from frozen sections of passaged tumors for mutational analysis by Signo.
[0228] Table 3. Source information for primary human tumors
[0229]
[0230] Xenografts whose name begins with LXF were obtained from Oncotest GmbH, Freiburg im Breisgau, Ge...
example 2
[0255] Example 2. Glucose Transporter
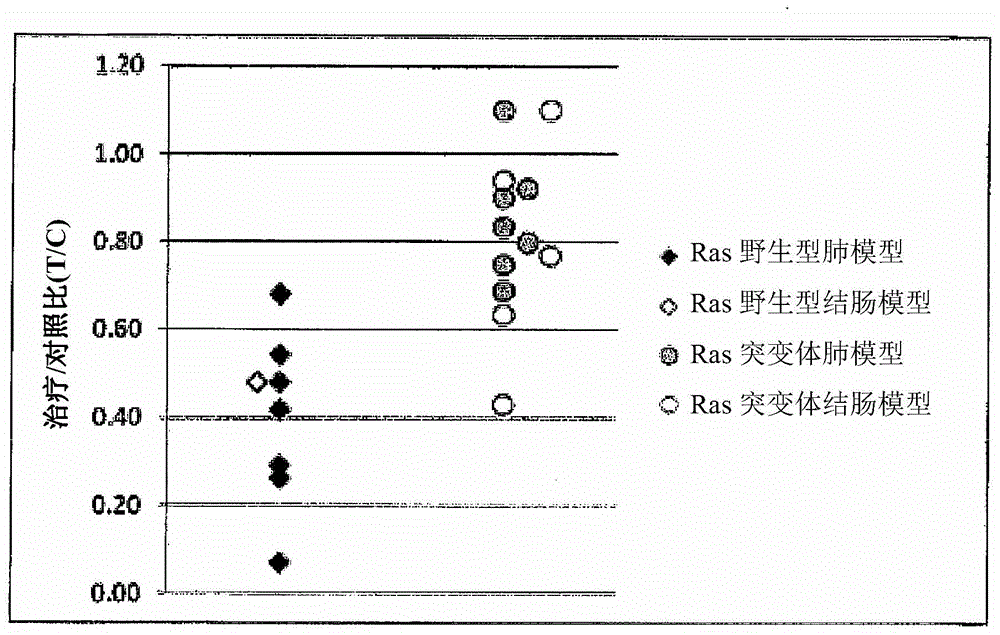
[0256] In general, tumor cells exhibit higher levels of glucose metabolism than normal cells (reviewed in Adkula et al. (2012) 24:650-654). KRAS mutant colorectal cancer cells display higher glucose uptake and glycolysis and better growth and survival under nutrient stress than wild-type cells (Yun et al. 2009 Science 325:1555). The study identified GLUT1 (glucose transporter 1) as upregulated in KRAS mutant colorectal cancer cells.
[0257] Most tumor cell lines are sensitive to MLN2238 in vitro under standard culture conditions using high glucose, making it difficult to distinguish between sensitive and resistant cell lines in vitro. However, under in vivo conditions, less glucose is available. Glucose transporter expression was analyzed by Western blotting of tumor cell lines listed in Table 5 that were not treated with MLN2238 but grown in mice as xenografts or in culture. Proteins were prepared from xenograft tumor samples or c...
example 3
[0258] Example 3. Other Sequencing Methods and General Procedures
[0259] Sanger sequencing method. PCR amplification was performed using optimized cycling conditions on a gene-to-exon basis. Primer extension sequencing was performed using Applied Biosystems BigDye version 3.1. Reactions were then run on an Applied Biosystems 3730xl DNA Analyzer. Sequencing base calling was performed using KBTM base caller (Applied Biosystems). Somatic mutation calls were determined by Mutation Measurer (SoftGenetics) and manually confirmed by aligning the sequencing data to the corresponding reference sequence using Seqman (DNASTAR).
[0260] Next-generation sequencing (NGS) methods. Directed NGS using the Illumina platform (Illumina Inc., San Diego, CA) was used to confirm and identify low frequency mutations in the markers. Primer pairs were designed to amplify coding exons. PCR products were quantified using PicoGreen assay and combined in equimolar ratios for each sample. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com