SSR DNA markers for jute expressed sequence tags
A technology for expressing sequence tags and DNA markers, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, microbial determination/inspection, etc., and can solve the problems of limited research on jute microsatellites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1:Sixty-six new microsatellite markers suitable for the analysis of jute genetic diversity were developed. The 66 microsatellite marker numbers are respectively: CcSSR001-CcSSR045 (Jute EST-SSR), CoSSR046-CoSSR066 (Jute EST-SSR), as shown in Table 1.
Embodiment 2
[0026] Example 2: Ten polymorphic microsatellite markers CcSSR008, CcSSR013, CcSSR016, CcSSR024, CcSSR029, CcSSR040, CoSSR049, CoSSR050, CoSSR052 and CoSSR053 were identified. as table 3 and figure 1 shown.
[0027] Table 3 Polymorphism and amplification efficiency of jute EST-SSR
[0028]
[0029]
[0030]
[0031] ABCD indicates the level of clarity of the electrophoretic band pattern.
Embodiment 3
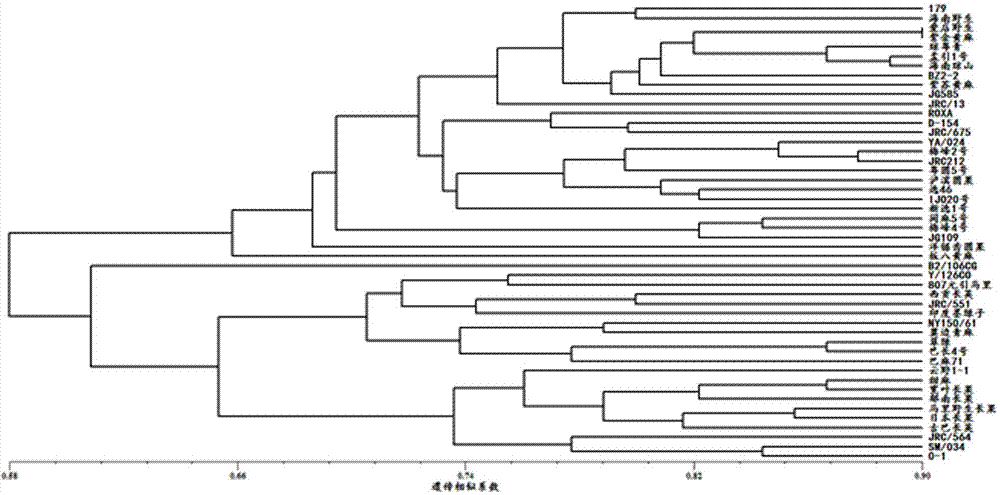
[0032] Example 3: Genetic diversity analysis of the genus Jute was analyzed using the microsatellite markers developed above. The test materials were 48 pieces of jute long-fruited wild species jute, long-fruited wild species jute, long-fruited cultivated species, round-fruited cultivated species jute and round-fruited wild species jute from 11 countries and regions (Table 4). . Using NTsys software to calculate the genetic similarity coefficient of 48 jute materials. The calculation method is unweighted cluster analysis (UPGMA), and the reading band adopts the method of 0 and 1, 0 means no band, and 1 means band. The results of cluster analysis showed that these 10 microsatellite markers could be well used in the genetic diversity analysis of jute. The results of cluster analysis are as follows: figure 2 As shown, 48 jute materials are divided into two categories, one is round fruit species and the other is long fruit species.
[0033] Table 4 Variety names and sourc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com