System and method for aligning genome sequence
A base sequence and sequence technology, which is applied in the field of analyzing the base sequence of the genome, to achieve the effects of improving accuracy, increasing the speed of global comparison, and improving the accuracy of mapping
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
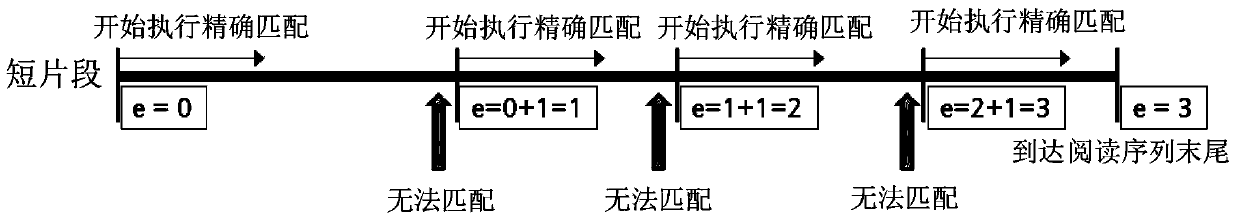
[0099] Example 1: When L=75, e=3,
[0100] Since f=15~15, so f=15,
[0101] s=4,
[0102] H=75–3×15–2×4=22.
example 2
[0103] Example 2: When L=100, e=4,
[0104] Since f=15~16, so f=16,
[0105] S=4,
[0106] H=100–4×16–2×4=36–8=28.
example 3
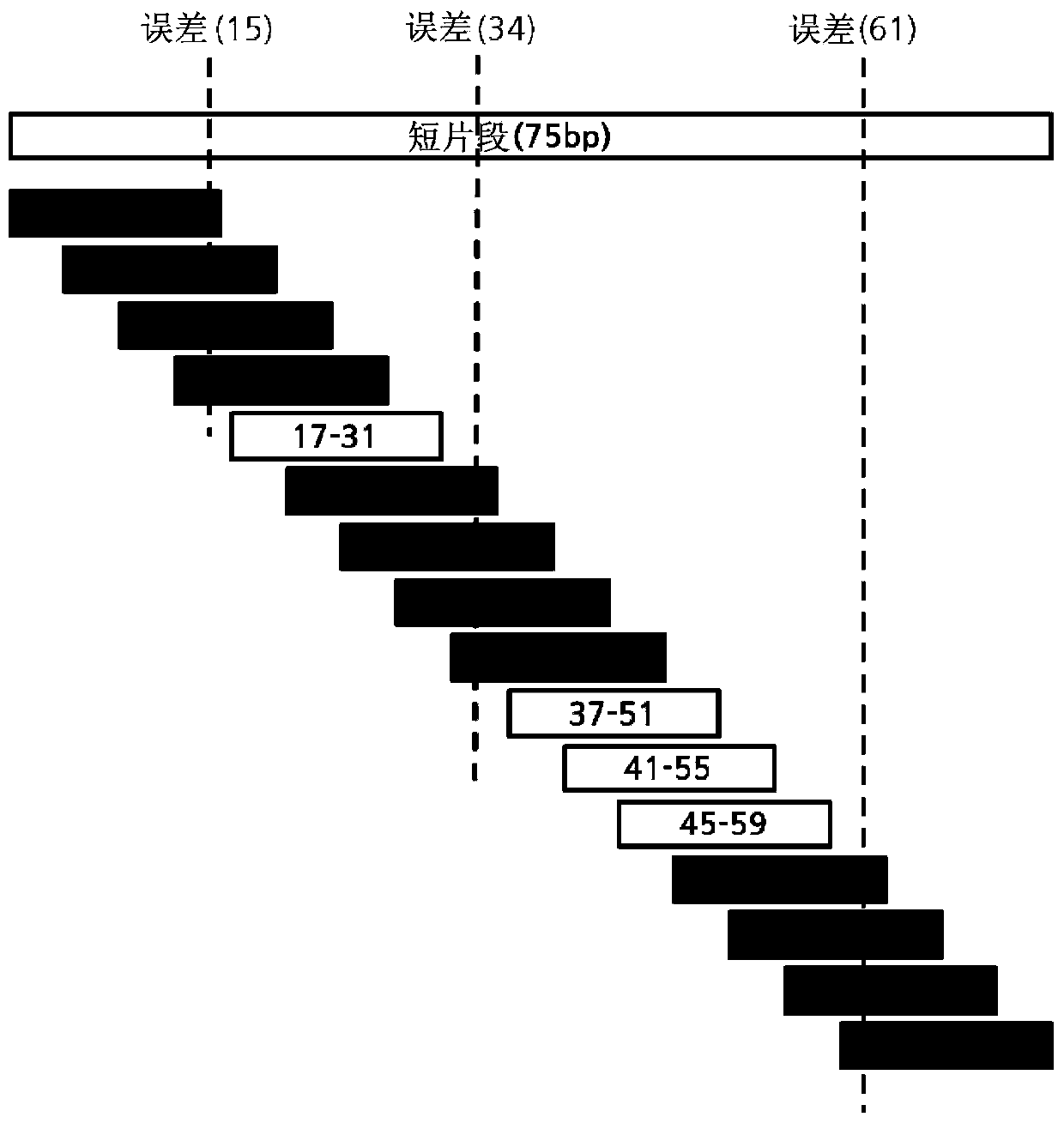
[0107] Example 3: When L=75, e=4,
[0108] Although f=15~12, since f should be greater than or equal to 15, so f=15,
[0109] s=4,
[0110] Although H=75–4×15–2×4=15-8=7, since f+s=19, the result will be H=19.
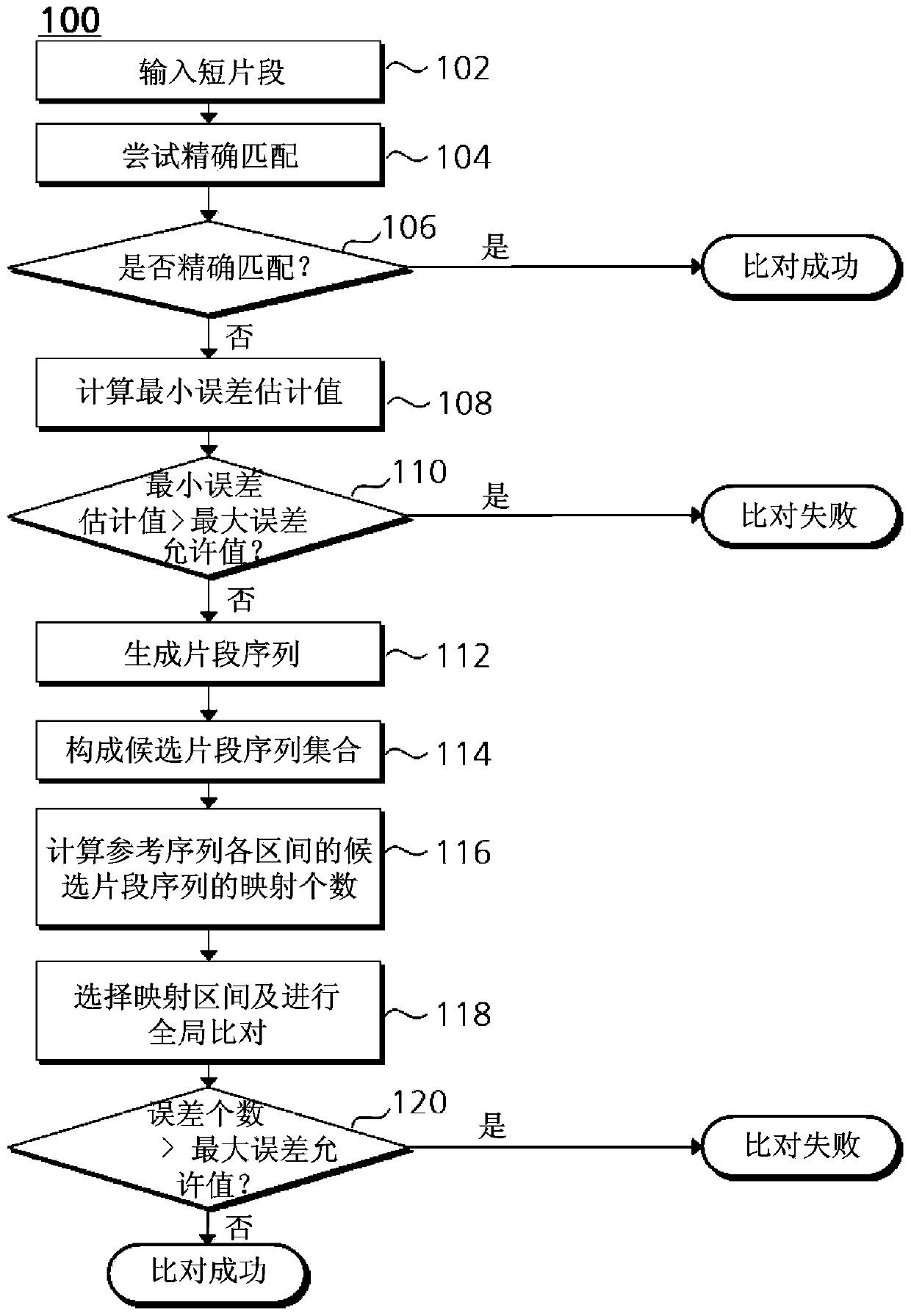
[0111] Figure 6 It is a block diagram of a base sequence alignment system 600 according to an embodiment of the present invention. The base sequence comparison system 600 according to one embodiment of the present invention is a device for performing the aforementioned base sequence comparison method, including: a fragment sequence generation unit 602, a screening unit 604, a mapping number calculation unit 606, and a comparison unit 608. The fragment sequence amplification unit 610.
[0112] The fragment sequence generation unit 602 generates a plurality of fragment sequences from short fragments obtained by a genome sequencer. As mentioned above, the fragment sequence generation unit 602 moves from the first base of the short fragment at a set interval and read...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com