Lung carcinoma cell specificity conjugation oligopeptide and application thereof
A lung cancer cell and specific technology, applied in the field of protein peptides, can solve the problem of few reports related to screening lung cancer targeting ligands, and achieve the effect of small molecular weight, high specificity and good tissue penetration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Screening of Lung Cancer Specific Binding Polypeptides
[0023] In this example, a phage display random 12-peptide library is used for subtractive screening of polypeptides that specifically bind to lung cancer cells, and the specific steps are as follows:
[0024] After digesting human lung cancer cells (A549) and human lung normal cells (MRC-5) with trypsin, adjust the cell density, inoculate in a culture dish pre-coated with poly-lysine, and wait until the cells grow to 85%-90% Screening experiments were performed for fusion.
[0025] Take the above-mentioned A549 cells, culture them with serum-free DMEM, add bovine serum albumin BSA to block, then add 10 μl phage peptide library, after incubation for 1.5 h, pour to remove unbound phage, and shake upside down to remove the residual solution. Rinse 3 times with washing solution 0.1% (v / v) TBST buffer, add non-specific buffer 0.2M glycine-hydrochloric acid (pH2.1) 1ml, suck out the eluate, add 150μl 1M Tris-...
Embodiment 2
[0029] Example 2 The amount of phage after screening and amplification
[0030] In Example 1, the phage obtained in each round of screening was diluted 100 times with LB medium, and 10 μl of the diluted phage was mixed with 200 μl of E. After the LB top layer agar, quickly pour it on the LB solid plate containing IPTG / Xgal, overnight, count the number of spots on the plate where the phage can grow, and then multiply this number by the dilution factor to get the plaque forming unit (pfu) drops per 10 μl phage Spend.
[0031] Enrichment of phage polypeptides specifically bound to lung cancer cells: As shown in Table 1, the recovery rate of phage clones positive for A549 was increased by 100 times after three rounds of selection.
[0032] Table 1 The recovery rate of positive phage after three rounds of screening
[0033]
Embodiment 3
[0034] Embodiment 3ELISA identification phage polypeptide
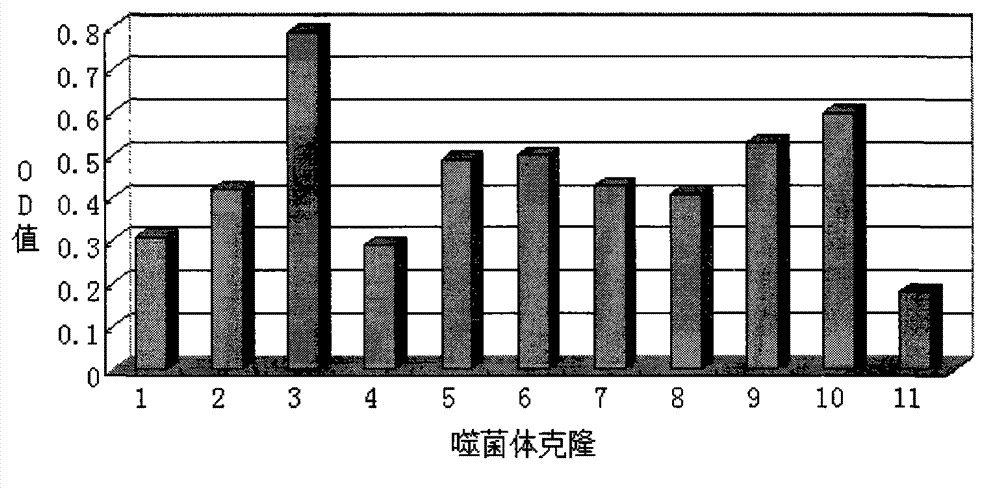
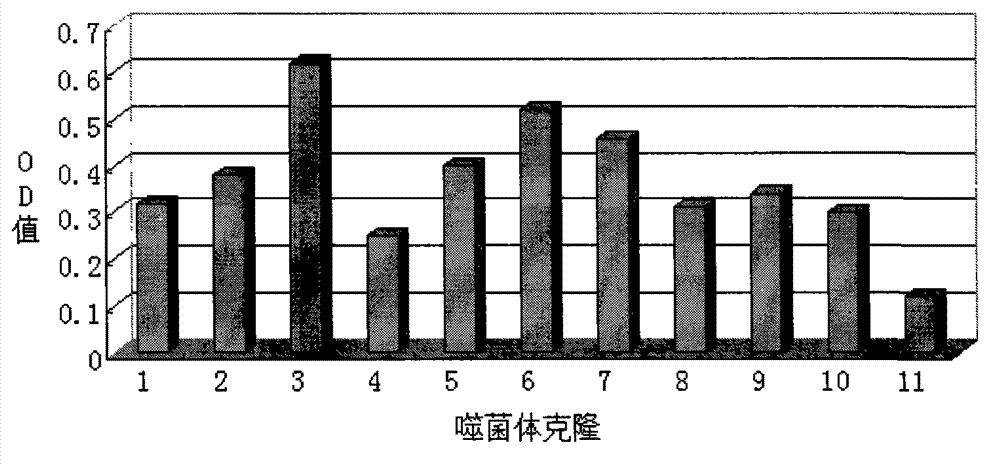
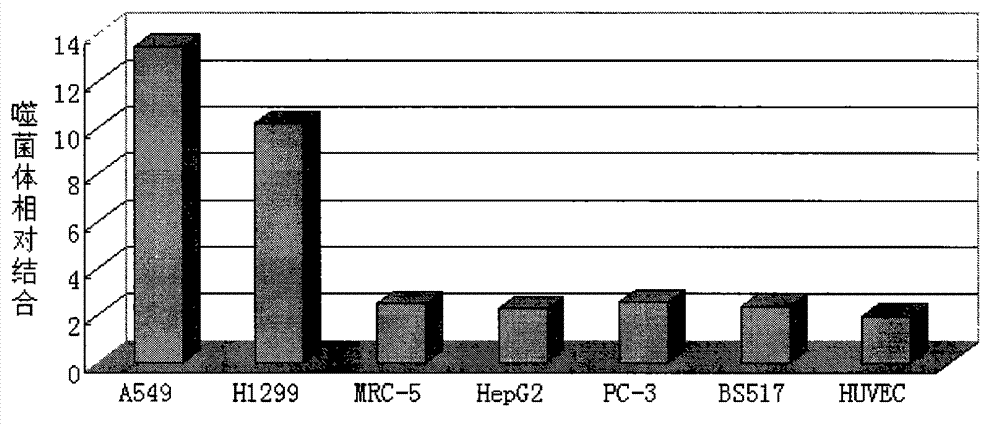
[0035] Identification of positive clones of phage polypeptides specifically binding to lung cancer: In Example 1, after three consecutive rounds of subtractive screening of the phage peptide library, 12 phage clones were randomly selected, and the phage clones were preliminarily identified for A549, A549, Affinity for H1299.
[0036] Press A549, H1299 by 1×10 5 The density per well was seeded in a 96-well plate and placed in CO 2 After culturing in the incubator for 24 hours, the cells were treated with serum-free for 1 hour, washed, fixed with paraformaldehyde, washed with PBS, treated with Triton X-100, blocked with PBS-BSA, added phage monoclonal, incubated for 2 hours; added HRP- Anti M13 antibody, incubate at 37°C for 1 h; use TMB for color development (50 μl TMB per well, color development for 10-15 minutes), add an equal volume of 1N HCl or 2N H 2 SO 4To terminate the reaction, the reaction solution in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com