Parallel gene splicing algorithm based on cluster map structure
A technology of gene splicing and graph structure, applied in the field of bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] A parallel gene mosaic algorithm based on the cluster graph structure, which includes creating a cluster graph and building a parallel framework;
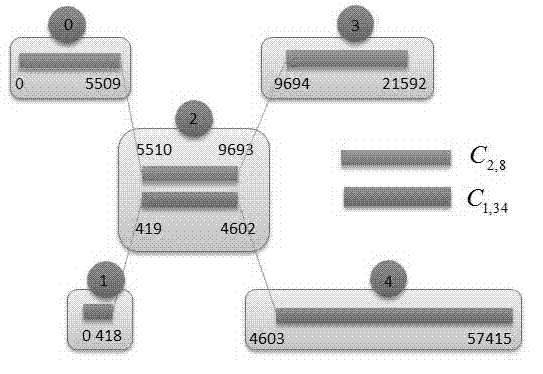
[0068] The creation of the cluster map refers to: according to the mapping results between the original gene data (short read length) and the long sequence (scaffold) generated by other algorithms, the similarity and matching degree of the scaffold are calculated, and then the clustering is performed. Two matching scaffolds constitute a scaffold pair (scaffold-pair), and all scaffold-pairs have multiple matching areas, and these areas are used as nodes, and the connections between them form edges to create a cluster graph;
[0069] Building a parallel framework refers to: running through the steps of the entire gene splicing algorithm, including reading and writing files, building indexes, short-read mapping, scaffold clustering, building cluster graphs, and search paths; The tasks in each step are divided, executed, and mer...
Embodiment 2
[0087] A parallel gene splicing algorithm based on the cluster graph structure proposed by the present invention can run on multiple operating systems (Linux, Mac, Windows), and the running method is very simple. The specific mode of operation of the program includes the following steps:
[0088] (1) Install all software package dependencies in the claims on the operating system;
[0089] (2) Prepare two types of data, the first data is the short sequence of the original paired-end sequencing gene, and the second is the output (long sequence) obtained by using data one as the input of multiple other gene splicing algorithms;
[0090] (3) Modify the path and parameters in the config.cfg file;
[0091] #------------input----------------
[0092] #########Mapping reads#####
[0093] Kmer_Size=30
[0094] Available_Processor_Num=20
[0095]Read_1= / home / ub / genome / realdata / SRR034959 / fasta / SRR034959_1.fasta
[0096] Read_2= / home / ub / genome / realdata / SRR034959 / fasta / SRR034959_2.fa...
Embodiment 3
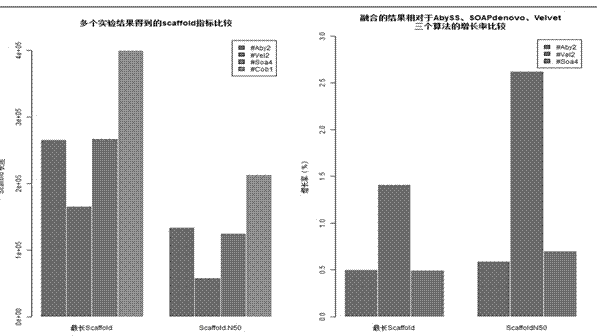
[0121] The following table is the method of the present invention and the existing three conventional gene splicing algorithms (ABySS, Velvet, SOAPdenove)
[0122] In Escherichia coli K-12 MG1655 (NCBI SRA accession
[0123] ERR022075, http: / / trace.ncbi.nlm.nih.gov / Traces / sra / sra.cgiview=run_browser&run=ERR022075) Comparison of the results of the data set, where #Aby indicates the experiment number of the algorithm ABySS, #Vel indicates the algorithm Velvet #Soa represents the experiment number of the algorithm SOAPdenove, and #Cob represents the experiment number of the present invention, obviously the advantages of the present invention are very obvious.
[0124]
[0125] in conclusion:
[0126] (1) The method of the present invention greatly increases the length of the scaffold sequence. It has been tested that the percentage of the length of the longest sequence obtained on the E. coli gene data set is 50% higher than that of other conventional algorithms.
[0127] (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com