The specific primer sequence that can be applied in the method for identifying different fish and the dna molecular marker method for identifying different fish
A technology of DNA molecules and specific primers, used in recombinant DNA technology, DNA/RNA fragments, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
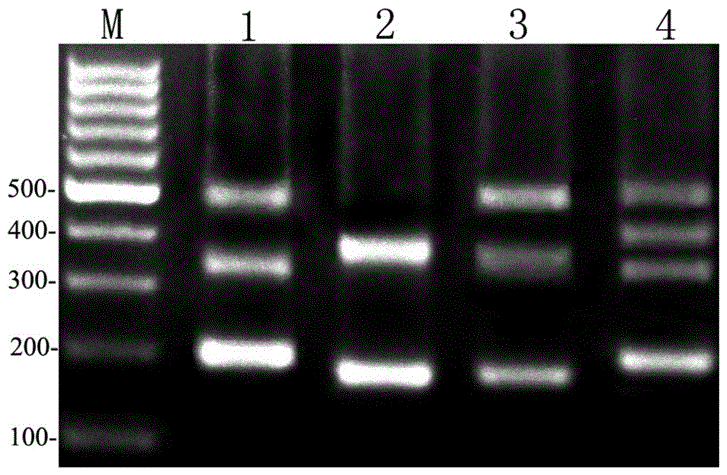
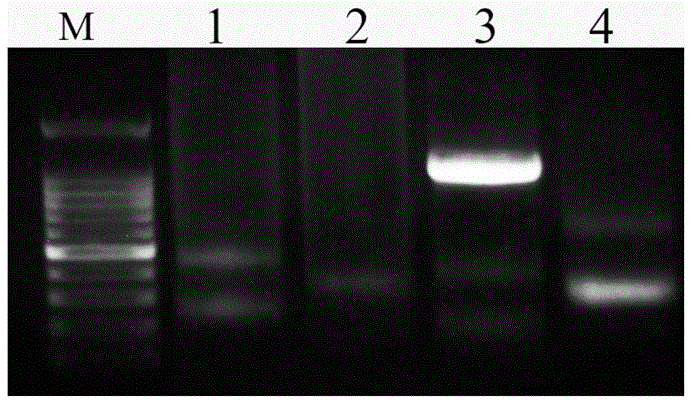
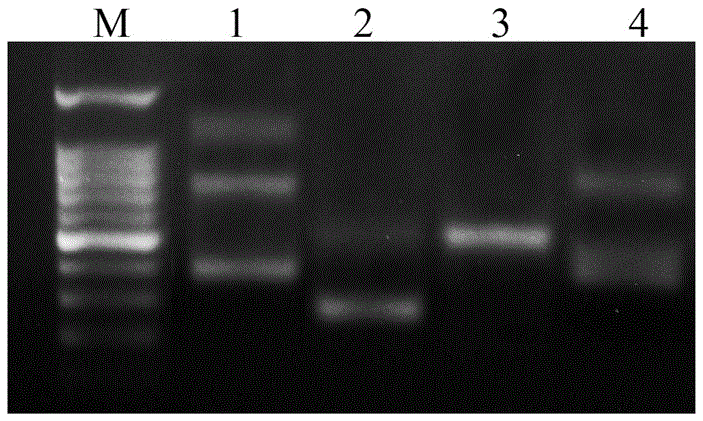
[0029] Referring to the known coding region sequences of 5S rDNA genes of other fishes, a specific primer sequence that can be used in the DNA molecular marker method for differentiating different fishes was designed with PrimerPremier5.0 software and Jellyfish1.4 software, provided by Shanghai Sangong Co., Ltd. Company (Shanghai Sangon for short) synthesized, the specific primer sequence is as follows:
[0030] Upstream primer: 5'-GCCCGATCTCGTCTGATCTCG-3';
[0031] Downstream primer: 5'-GCGCTCAGGTTGGTATGGCCG-3'.
[0032] A kind of DNA molecular marker method of different fishes with above-mentioned specific primer sequence identification of the present invention, comprises the following steps:
[0033]1. Prepare experimental materials.
[0034] Angelfish ( Pterophyllum ),carp( Cyprinus carpio L. ), yellow-tailed pomfret ( Xenocypris davidi Bleeker ), Crawfish ( Erythroculterilisha eformis Bleeker ), moray eel ( Muraenes oxcinereus ), Japanese eel ( Anguilla Japo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com