Enzyme-responsive self-aggregation luminous molecule and applications thereof in monitoring enzyme activity
A self-aggregation luminescent, enzyme-responsive technology, applied in the direction of chemiluminescence/bioluminescence, and analysis by making materials undergo chemical reactions, to achieve excellent biocompatibility, strong biocompatibility, and avoid aggregation-induced luminescence quenching and the effect of photoquenching
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
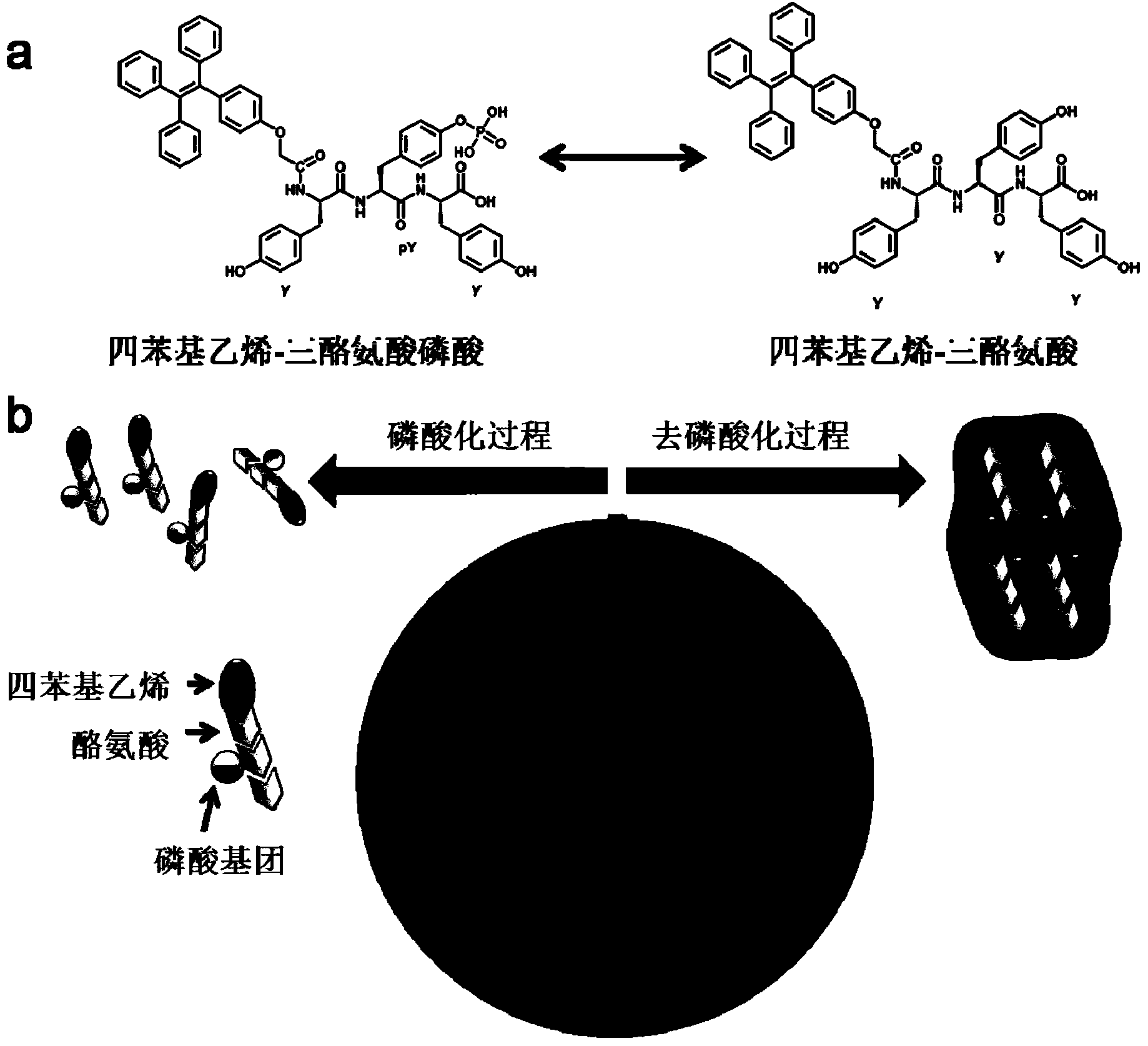
[0038] First, this example describes the synthesis of tetraphenylethylene carboxylic acid derivatives, a self-aggregating luminescent molecule, with the following route:
[0039]
[0040] (I) Preparation of I, I, 2-triphenyl-2-Gv-methoxyphenyl)ethene: 3.68g, 20mmol of benzophenone was added to a clean three-neck round bottom flask, and then 19.3g, 100mmol of zinc powder was added. The flask was evacuated and filled with dry nitrogen three times. Afterwards, inject 200mL of anhydrous tetrahydrofuran into the flask, add 6.8mL, 60mmol of titanium tetrachloride dropwise at 0°C, reflux overnight and add 10% potassium carbonate solution (13.8g of potassium carbonate dissolved in 125mL of water) to terminate the reaction. After the phases were separated, the organic phase was washed twice with brine and dried over anhydrous magnesium sulfate, then filtered and concentrated. The obtained crude product was purified by silica gel column, and was eluted with petroleum ether and dieth...
Embodiment 2
[0048] First, synthesize TPE with the same method and process as in Example 1.
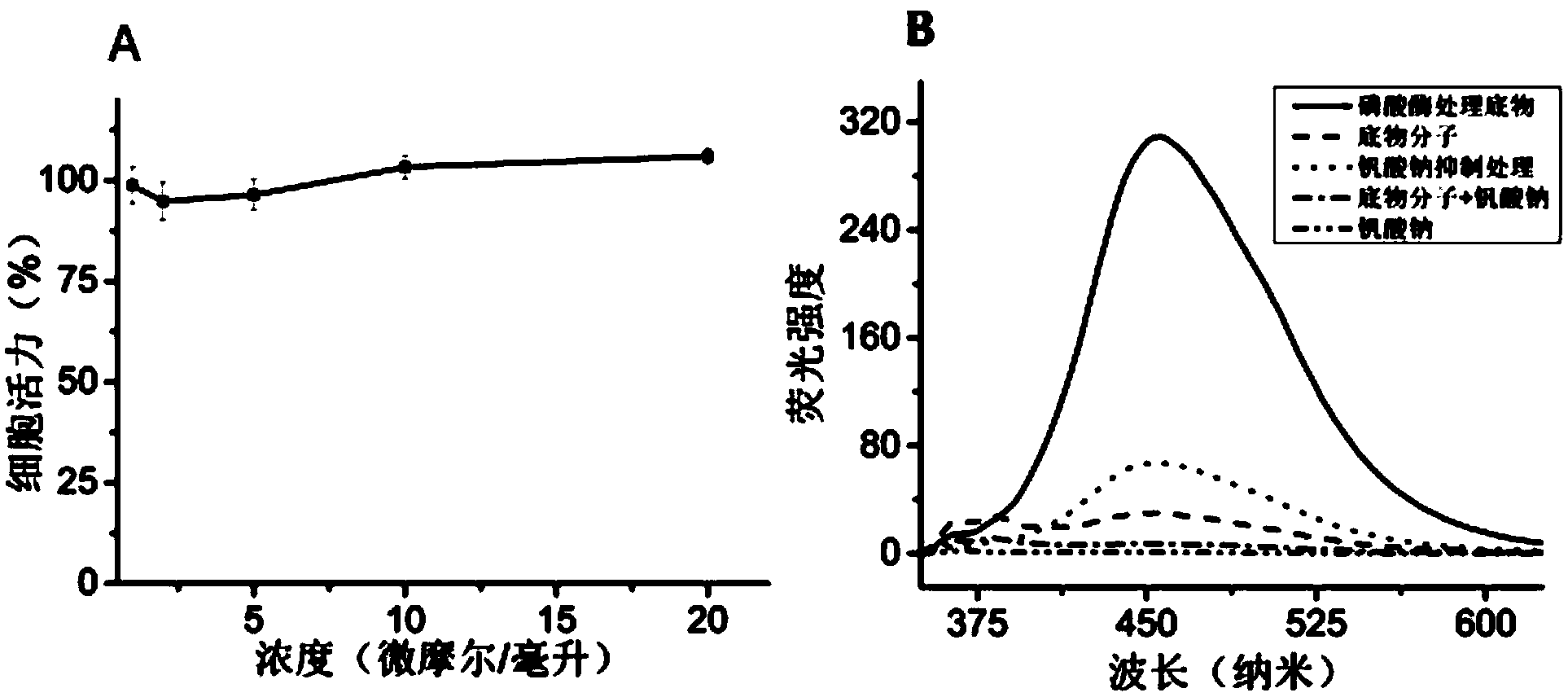
[0049] Secondly, in this embodiment, the live cell imaging of protein tyrosine phosphatase 1B (Protein Tyrosine Phosphatase 1B, PTP1B) activity in human Hela cells is carried out by including the following steps.
[0050] (1) The synthetic substrate sequence is: TPE-TPE-DADEpYL. The protected amino acids used and catalysts DIEA, HATU, DMAP, DCC and hexahydropyridine were purchased from Novabiochem, Acid Clear resin for synthesis was purchased from Peptide International, and DMF and DCM were purchased from Fisher. First, the resin is swelled with DMF for 10-60 minutes. At the same time, 100mg-1000mg of the first leucine was dissolved in anhydrous DCM, and then 10mg-250mg of DCC was added, and stirred in an ice bath for 1-30 minutes until a white precipitate formed. Evaporate the excess DCM with a rotary evaporator, dissolve it with 1-10mL DMF, and add it to the swollen resin to react for 10-60min...
Embodiment 3
[0054] First, synthesize TPE with the same method and process as in Example 1.
[0055] Secondly, in this embodiment, live cell imaging of protein phosphatase 2A (Protein phosphatase 2A, PP2A) activity in MCF-7 cells is performed by including the following steps.
[0056] (1) The synthetic substrate sequence is: TPE-RRREEEpTEEEAA. The protected amino acids used and catalysts DIEA, HATU, DMAP, DCC and hexahydropyridine were purchased from Novabiochem, Acid Clear resin for synthesis was purchased from Peptide International, and DMF and DCM were purchased from Fisher. First, the resin is swelled with DMF for 10-60 minutes. At the same time, 100mg-1000mg of the first alanine was dissolved in anhydrous DCM, and then 10mg-250mg of DCC was added, and stirred in an ice bath for 1-30 minutes until a white precipitate formed. Evaporate the excess DCM with a rotary evaporator, dissolve it with 1-10mL DMF, and add it to the swollen resin to react for 10-60min. Then wash the resin with ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com