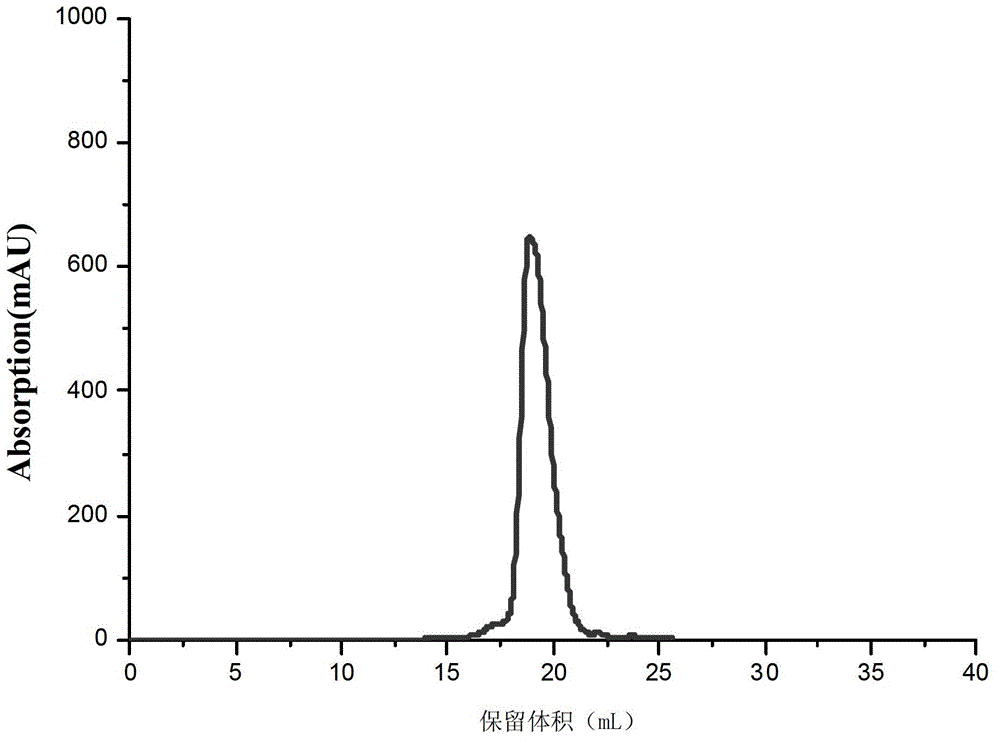
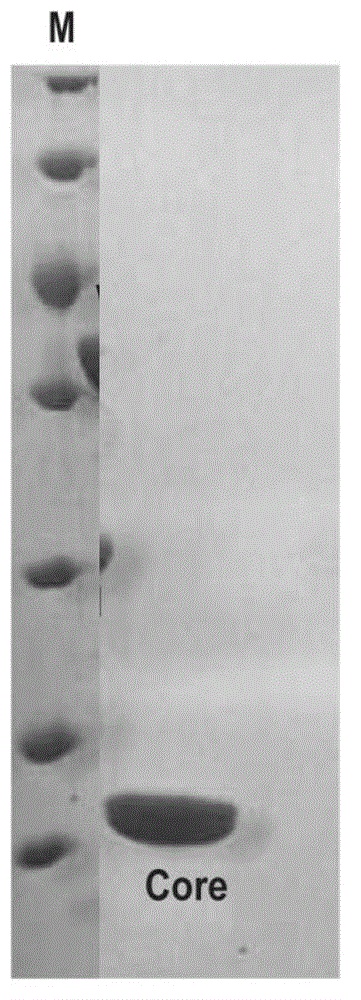
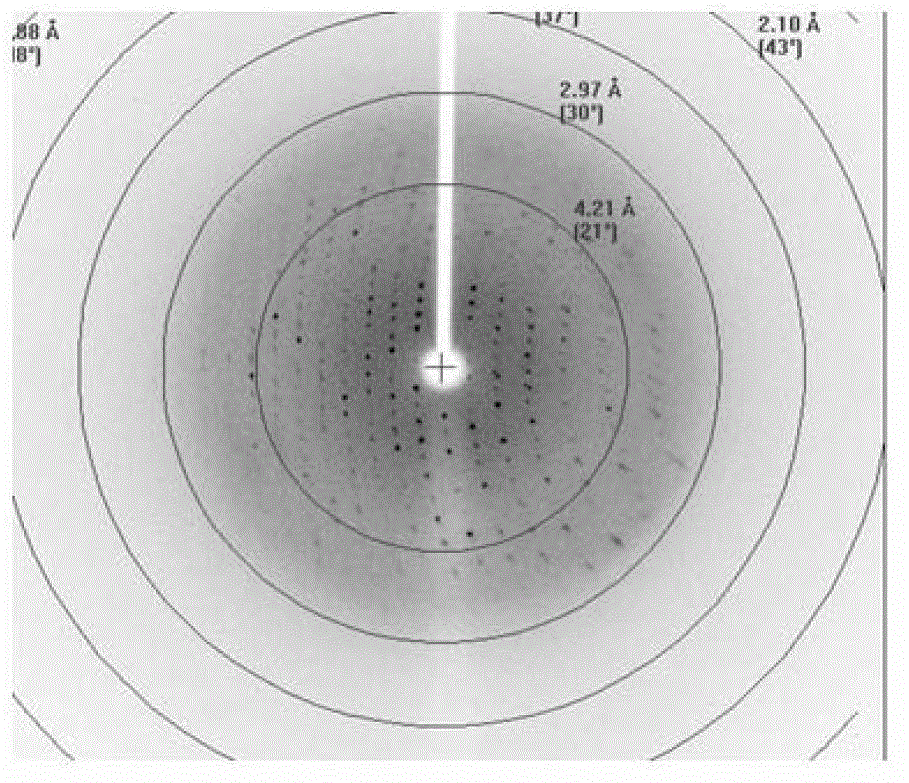
Core structural domain of VirB protein, and coding genes and applications thereof
A protein and residue technology, applied in applications, genetic engineering, plant genetic improvement, etc., can solve problems such as bacterial metabolic process burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, the preparation of VirB core fragment
[0039] 1. Construction of recombinant plasmids
[0040] Insert the double-stranded DNA molecule shown in the 64th to 429th nucleotides of the sequence 2 of the sequence listing from the 5' end between the Nde1 and Xhol restriction sites of the plasmid pET-28a(+), to obtain the recombinant plasmid pET28a-VirB core. In the recombinant plasmid pET28a-VirB core, the foreign gene and some nucleotides in the plasmid pET-28a (+) form the fusion gene shown in the sequence 2 of the sequence table, and express the fusion protein shown in the sequence 1 of the sequence table (sequence In 1, the 5th to 10th amino acid residues from the N-terminal form the His tag, and the 22nd to 143rd amino acid residues form the VirB core fragment)
[0041] 2. Preparation of VirB core fragments
[0042] 1. Introduce the recombinant plasmid pET28a-VirB core obtained in step 1 into competent cells of Escherichia coli Rosseta to obtain recombi...
Embodiment 2
[0053] Embodiment 2, the preparation of Selenium VirB core fragment
[0054] 1. Transform Escherichia coli B834 with the recombinant plasmid pET28a-VirB core obtained in step 1 of Example 1 to obtain recombinant bacteria.
[0055] 2. Inoculate the single clone of the recombinant bacteria obtained in step 1 into 3 ml of LB liquid medium containing 50 μg / μL kanamycin, and cultivate overnight at 37° C. and 250 rpm with shaking.
[0056] 3. Take the bacterial liquid obtained in step 2, inoculate it into 200 mL of LB liquid medium containing 50 μg / μL kanamycin, incubate at 37°C and 250 rpm for 12 hours, and collect the bacterial precipitate by centrifugation.
[0057] 4. Preparation of selenomethionine medium: (1) Weigh 86.4 grams of medium base from the reagent kit (Molecular dimensions company, product number MD12-501B), dissolve it in 4L of ultrapure water, and autoclave at 121°C for 15 minutes , stored at 4°C after cooling; (2) Weigh 20.4 grams of nutrient mix (Molecular dimen...
Embodiment 3
[0062] Example 3, Crystal Analysis of the Complex Formed by VirB core Fragment and DNA Molecule
[0063] 1. Assembly of the complex
[0064] 1. Synthesize single-stranded DNA molecule A and single-stranded molecule B respectively, dissolve each single-stranded DNA molecule in pH 8.5, 100mM Tris buffer and make the concentration 100μM, and then mix the two single-stranded DNA molecules Equal volumes of the solutions were mixed, incubated at 95°C for 10 minutes, and naturally cooled to 4°C to obtain a double-stranded DNA molecule solution (icsB solution).
[0065] Single-stranded DNA molecule A: 5'-AAA CTCGTTTCATCATGAAATCCCAC-3';
[0066] Single-stranded DNA molecule B: 3'-GAGCAAAGTAGTACTTTAGGGTG TTT-5'.
[0067] 2. Mix the VirB core fragment solution obtained in Example 1 and the double-stranded DNA molecule solution obtained in step 1 according to the equimolar ratio of protein and DNA, and incubate on ice for 3 hours.
[0068] 3. Synthesize single-stranded DNA molecule C a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| fatigue bending times | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com