Core structural domain of VirB protein, and coding genes and applications thereof
A gene and encoding technology, applied to the core domain of VirB protein and its encoding gene and application field, can solve the problem of bacterial metabolic process burden and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
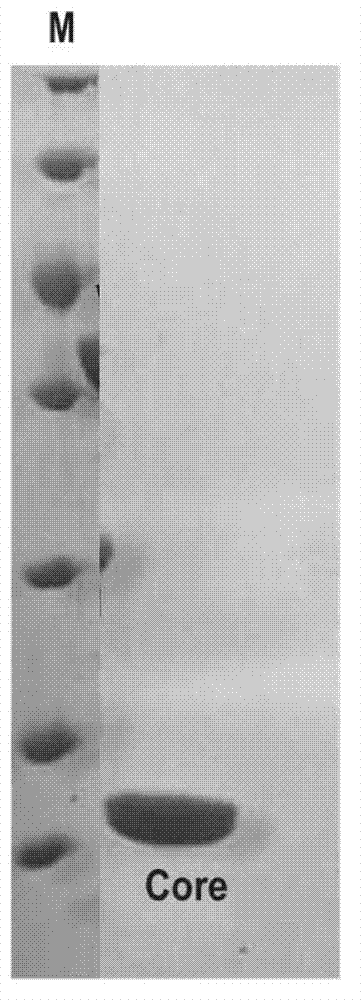
[0038] Example 1. Preparation of VirB core fragments
[0039] 1. Construction of recombinant plasmid
[0040] Insert the double-stranded DNA molecule shown in sequence 2 from the 64th to 429th nucleotides of the 5'end of the sequence table between the Nde1 and Xhol restriction sites of plasmid pET-28a(+) to obtain the recombinant plasmid pET28a-VirB core. In the recombinant plasmid pET28a-VirB core, the foreign gene and some of the nucleotides in the plasmid pET-28a(+) form the fusion gene shown in sequence 2 of the sequence list, and express the fusion protein shown in sequence 1 of the sequence list (sequence In 1, the 5th to 10th amino acid residues from the N-terminal form the His tag, and the 22nd to 143th amino acid residues form the VirB core fragment)
[0041] 2. Preparation of VirB core fragments
[0042] 1. The recombinant plasmid pET28a-VirB core obtained in step 1 is introduced into competent cells of Escherichia coli Rosseta to obtain recombinant bacteria.
[0043] 2. In...
Embodiment 2
[0053] Example 2. Preparation of VirB core fragments
[0054] 1. Transform the recombinant plasmid pET28a-VirB core obtained in step 1 of Example 1 into Escherichia coli B834 to obtain recombinant bacteria.
[0055] 2. Inoculate the single clone of the recombinant bacteria obtained in step 1 into 3ml of LB liquid medium containing 50μg / μL kanamycin, and cultivate overnight with shaking at 37°C and 250rpm.
[0056] 3. Take the bacterial solution obtained in step 2 and inoculate it into 200 mL of LB liquid medium containing 50 μg / μL kanamycin, incubate at 37° C. and 250 rpm for 12 hours, and collect the bacterial pellet by centrifugation.
[0057] 4. Preparation of selenomethionine medium: (1) Weigh 86.4 grams of medium base from the reagent kit (Molecular Dimensions Company, Catalog No. MD12-501B), dissolve it in 4L ultrapure water, and autoclave at 121°C for 15 minutes , Keep it at 4℃ after cooling; (2) Weigh 20.4g of nutrient mix (Molecular Dimensions Company, Product No. MD12-502) f...
Embodiment 3
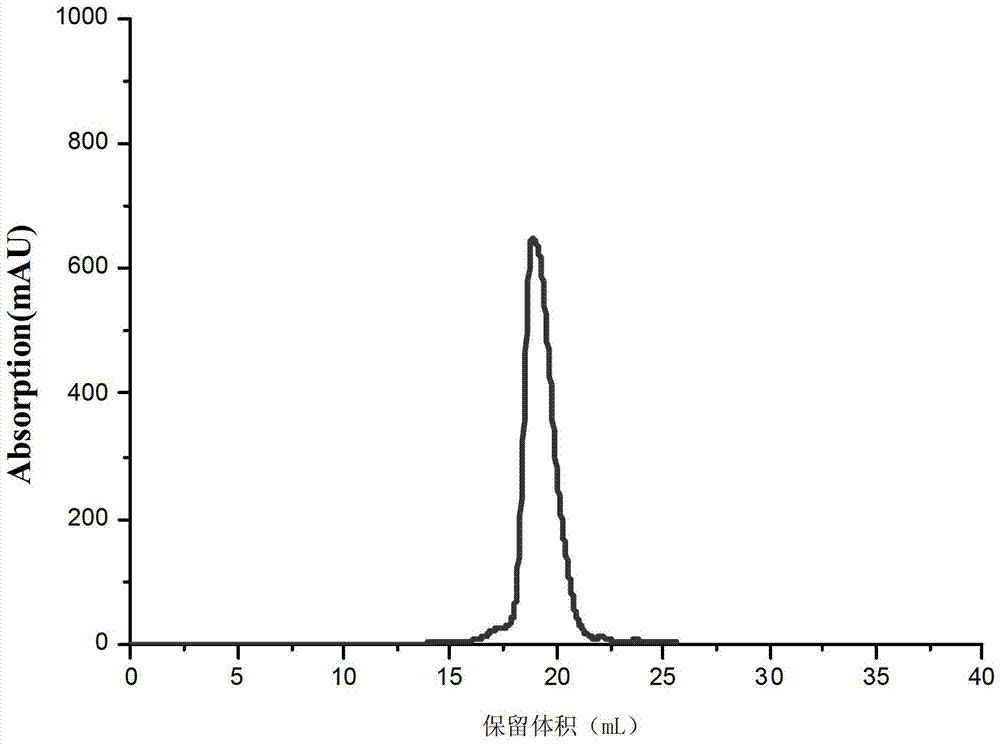
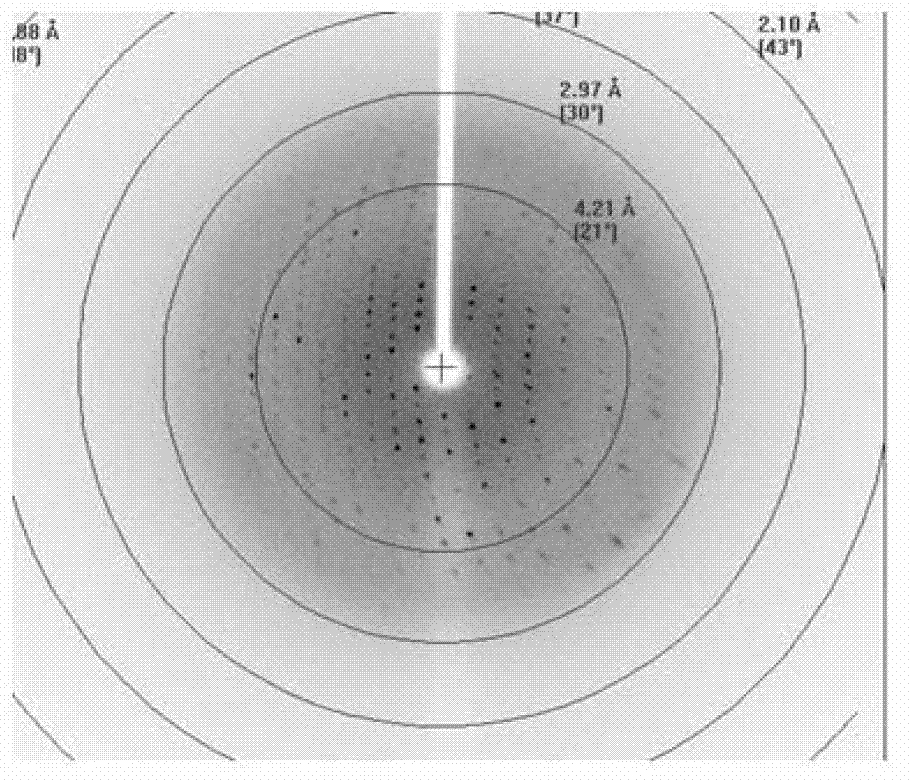
[0062] Example 3. Crystal analysis of the complex formed by VirB core fragments and DNA molecules
[0063] 1. The assembly of the complex
[0064] 1. Synthesize single-stranded DNA molecule A and single-stranded molecule B separately. Dissolve each single-stranded DNA molecule in a pH 8.5, 100 mM Tris buffer and make the concentration 100 μM, and then combine the two single-stranded DNA molecules The solutions were mixed in equal volumes, incubated at 95°C for 10 minutes, and naturally cooled to 4°C to obtain a double-stranded DNA molecule solution (icsB solution).
[0065] Single-stranded DNA molecule A: 5’-AAA CTCGTTTCATCATGAAATCCCAC-3’;
[0066] Single-stranded DNA molecule B: 3'-GAGCAAAGTAGTACTTTAGGGTG TTT-5'.
[0067] 2. Mix the VirB core fragment solution obtained in Example 1 and the double-stranded DNA molecule solution obtained in Step 1 according to an equimolar ratio of protein and DNA, and incubate on ice for 3 hours.
[0068] 3. Synthesize single-stranded DNA molecule C and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| fatigue bending times | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com