Fluorescent PCR method to identify drug-resistant mutations of Campylobacter jejuni and macrolides
A macrolide, Campylobacter jejuni technology, applied in fluorescence/phosphorescence, biochemical equipment and methods, microbial determination/inspection, etc. and other problems, to achieve the effect of high resolution, increased hybridization stability, and accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Embodiment 1: Design and performance investigation of primers and probes
[0082] According to the VS1 gene sequence (GI: 296939), 23S rRNA sequence (GI: 3245050) and rplD gene sequence (GI: 905980) of the whole genome (NC_002163.1) of the standard strain of Campylobacter jejuni (Campylobacter jejuni NCTC11168=ATCC700819) published by NCBI , using Primer Premier 5 and Primer Express 2.0 software to design three pairs of primers, which were used to amplify the 214bp fragment of the VS1 gene unique to Campylobacter jejuni (position 931-1144 of the VS1 whole gene), the 96bp fragment of the 23S rRNA gene (the 23S rRNA whole gene 2020-2115) and a 134bp fragment of the rplD gene (120-253 of the rplD gene). Primer Premier 5 and Primer Express 2.0 software were used to evaluate the designed primers, and the primers with the strongest binding ability and the lowest probability of dimer formation were selected. In addition, the BLAST tool in NCBI was used to evaluate the specifi...
Embodiment 2
[0086] Embodiment 2, the construction of seven kinds of control plasmids
[0087] 1. Construction of wild-type 23S rDNA and rplD control plasmids
[0088] According to the 23S rRNA sequence (GI: 3245050) and the rplD gene sequence (GI: 905980) of the whole genome (NC_002163.1) of the standard strain of Campylobacter jejuni (Campylobacter jejuni NCTC11168=ATCC700819) published by NCBI, use Primer Premier 5 and Primer Express 2.0 The software designed two pairs of primers, which were used to amplify the 147bp fragment at positions 1995-2141 of the unique 23S rRNA gene of Campylobacter jejuni and the 270bp fragment at positions 120-389 of the rplD gene. location. The designed primers were entrusted to Nanjing GenScript Biotechnology Co., Ltd. for synthesis. The forward and reverse sequences of the two pairs of primers are shown in Table 2, and the binding base sequences of the two pairs of primers in the 23S rDNA and the rplD whole gene are shown in Table 2. figure 1 B and fi...
Embodiment 3
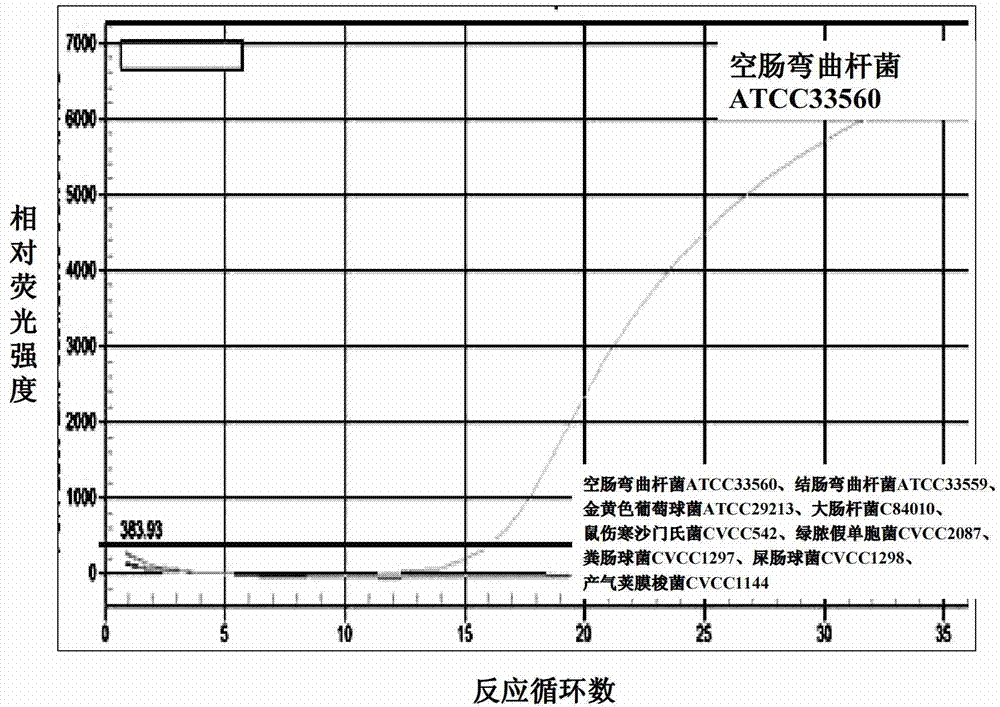
[0104] Embodiment 3: Optimization of multiple real-time fluorescent PCR reaction system and reaction conditions
[0105] Primers, probes, Mg in the multiplex real-time fluorescent PCR reaction system 2+ Concentration, concentration of Taq enzyme, etc. are the key parameters affecting the effect of fluorescent PCR amplification. In the present invention, 3 pairs of primers and 4 probes that have been designed and synthesized are placed in the same reaction system for multiple real-time fluorescent PCR, and the concentration of each component in the reaction system is optimized: the concentration of the primers is increased from 0.2 to 0.6uM by 0.05μM. The needle concentration is increased from 0.1μM to 0.45μM by 0.05μM, the concentration of dNTP is increased by 0.025mM from 0.075mM to 0.25mM, the concentration of Mg2+ is increased by 0.25mM from 0.75mM to 2.5mM, and the amount of Taq enzyme is increased by 0.5U from 1U to 5U . Only one variable is set for each test and the sa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com