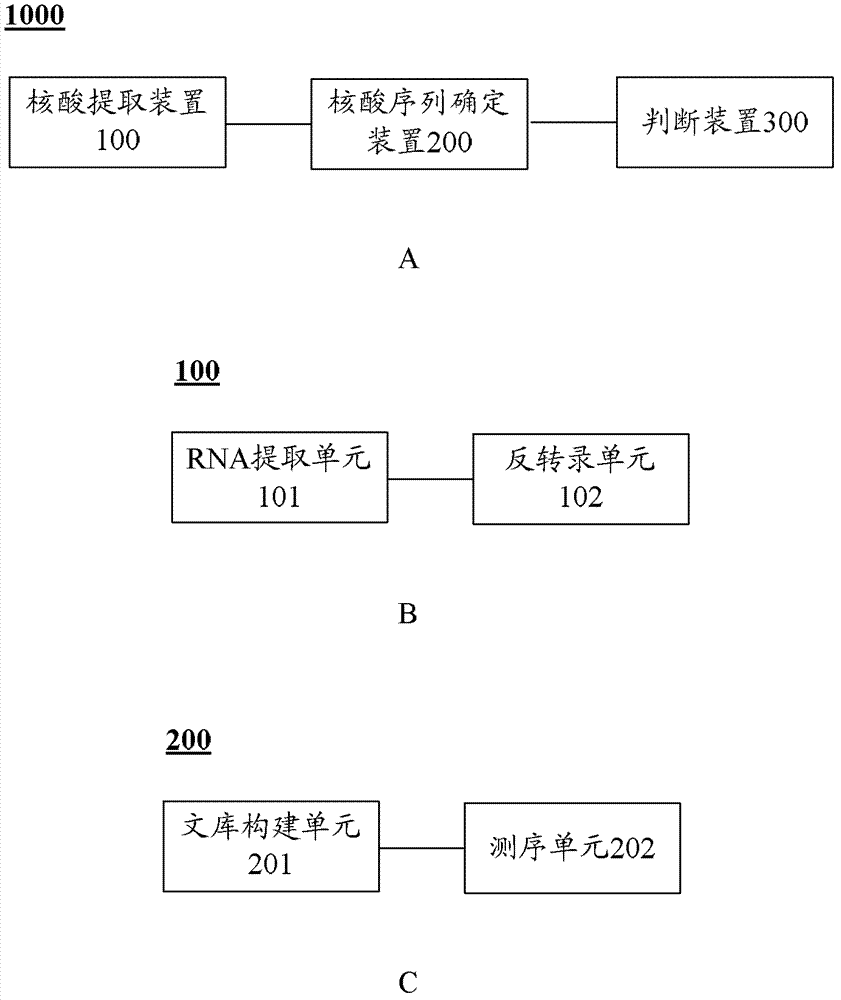
SNRNP200 gene mutant and application thereof
A technology for mutants and degenerative diseases, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve problems that need to be further studied
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0222] Example 1 High-throughput exome sequencing
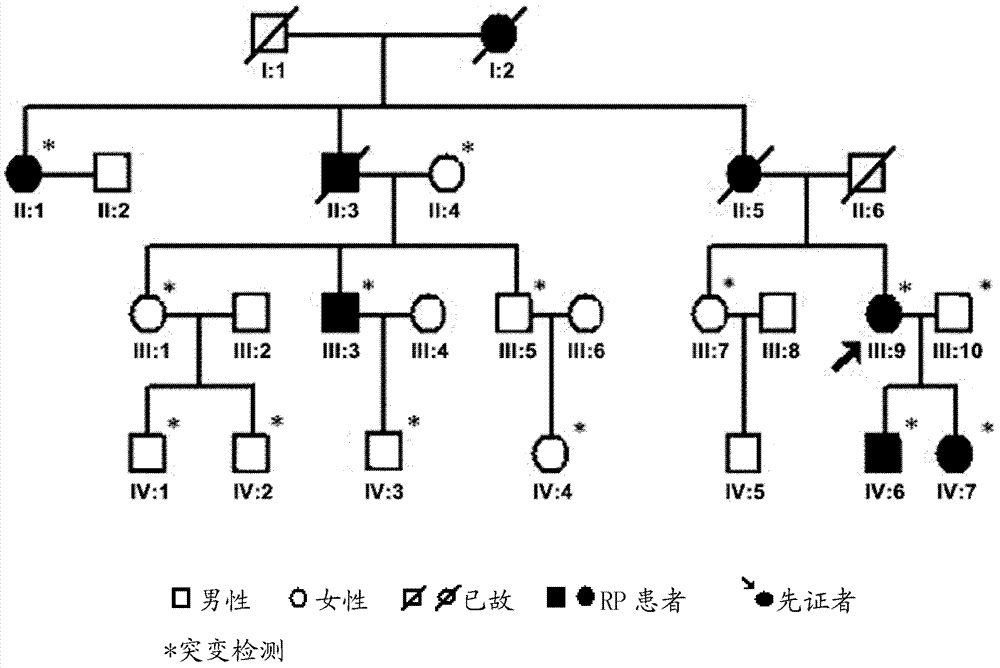
[0223] For the probands in the aforementioned ADRP family ( figure 2 III in: 9) Exome sequencing was performed to obtain candidate de novo SNP sites.
[0224] Specific steps are as follows:
[0225] 11 Genomic DNA preparation: collection of probands in the aforementioned ADRP family ( figure 2 In III: 9) peripheral blood, the genomic DNA in the peripheral blood leukocytes was extracted by the conventional phenol-chloroform method, and the concentration and purity of the DNA were measured by a spectrophotometer, and the OD260 / OD280 of each sample genomic DNA was located at 1.7 Between -2.0, the concentration is not less than 200ng / microliter, and the total amount is not less than 30 micrograms.
[0226] 1.2 High-throughput sequencing:
[0227] The DNA captured by the NimbleGen SeqCap EZ Human Exome Library v2.0 whole-exon capture platform was sequenced using Illumina Hiseq2000.
[0228] 1.3 Exome sequencing data analysis:...
Embodiment 2
[0230] Example 2 Sanger sequencing verification of candidate de novo SNP sites
[0231] Design primers for SNRNP200 (exon 20 2653C>G), PDE6B (exon 11 1415G>A), USH2A (exon 5997T>C), USH2A (exon 12 2750G>A) and USH2A (exon 51 10246T>G) sequences, by PCR Amplification, product purification, and sequencing methods to obtain relevant sequences. Sequencing by Sanger method confirmed that among the candidate de novo SNP sites obtained after high-throughput sequencing, SNRNP200 (exon 202653C>G), PDE6B (exon 111415G>A) and USH2A (exon 12 2750G>A) were de novo SNP sites , while USH2A (exon 51 10246T>G) and USH2A (exon 5 997T>C) were false positive sites.
[0232] 2.1 DNA extraction
[0233] Members in the aforementioned ADRP family were collected ( figure 2 II:1, II:4, III:1, III:3, III:5, III:7, III:9, III:10, IV:1, IV:2, IV:6, IV:7) Peripheral blood, the specific method is the same as step 1.1 in Example 1.
[0234] 2.2 Primer design
[0235] The PCR reaction primers were desi...
Embodiment 3d
[0244] Example 3 Sanger sequencing verification of de novo SNP sites in the family and in the normal population control group
[0245] respectively for figure 2 There are 5 RP patients (II: 1, III: 3, III: 9, IV: 6, IV: 7) and 7 normal members (II: 4, III: 1, III: 5, III: 7) in the family shown , III: 10, IV: 1, IV: 2) and 100 normal people outside the pedigree were tested. Among them, primers were designed for SNRNP200 (exon 202653C>G), PDE6B (exon 111415G>A) and USH2A (exon 122750G>A) sequences, and relevant sequences were obtained by PCR amplification, product purification and sequencing. Mutant type or wild type, verify the correlation between the above three heterozygous mutations and RP. The specific method steps are as follows:
[0246] 3.1 DNA extraction: respectively in the family ( figure 2 ) 5 RP patients (II: 1, III: 3, III: 9, IV: 6, IV: 7), 9 normal members (II: 4, III: 1, III: 5, III: 7, III: 10. IV:1, IV:2, IV:3, IV:4) and peripheral venous blood of 100 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com