Method and kit for detecting non-small cell lung cancer drive gene mutation spectrum, and application
A non-small cell lung cancer, gene-driven technology, applied in biochemical equipment and methods, microbial determination/examination, etc., can solve the problems of long waiting time for patients, unable to meet the gene mutation spectrum, difficult to obtain materials for advanced patients, etc. Achieve the effect of shortening the detection time, saving the amount of DNA used, and making the results easy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] (1) Primer design
[0068] (1) Design primers to amplify the exon segments of the seven driver genes of non-small cell lung cancer
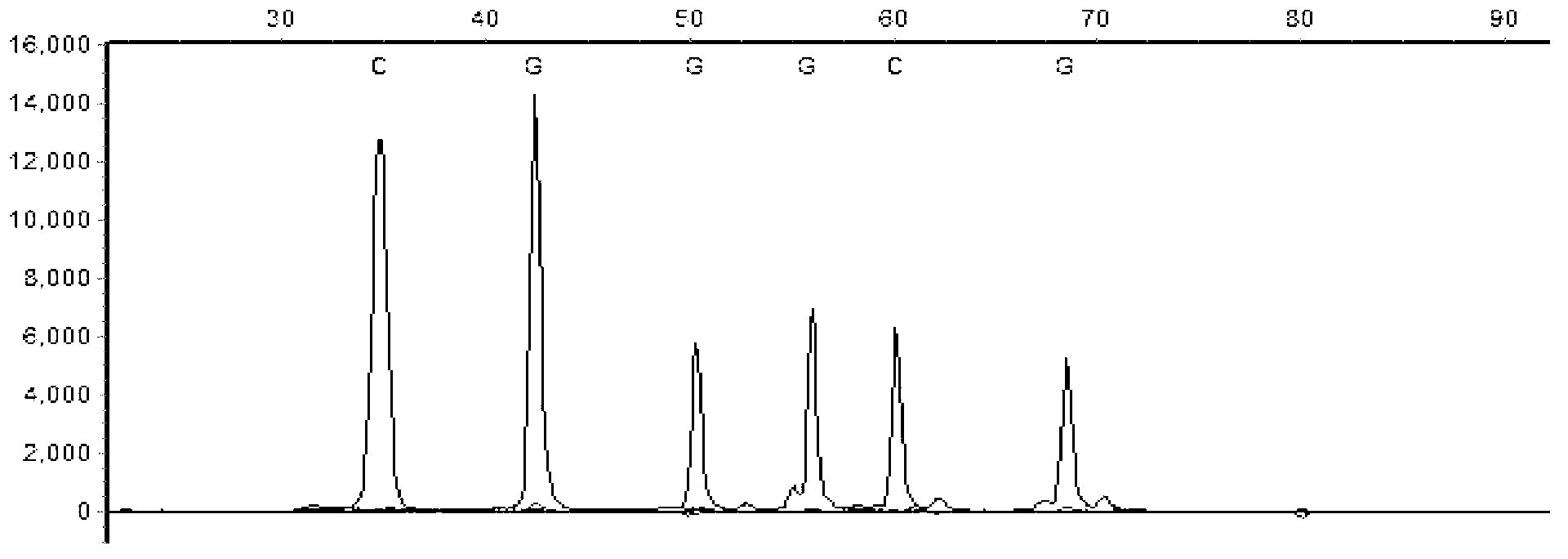
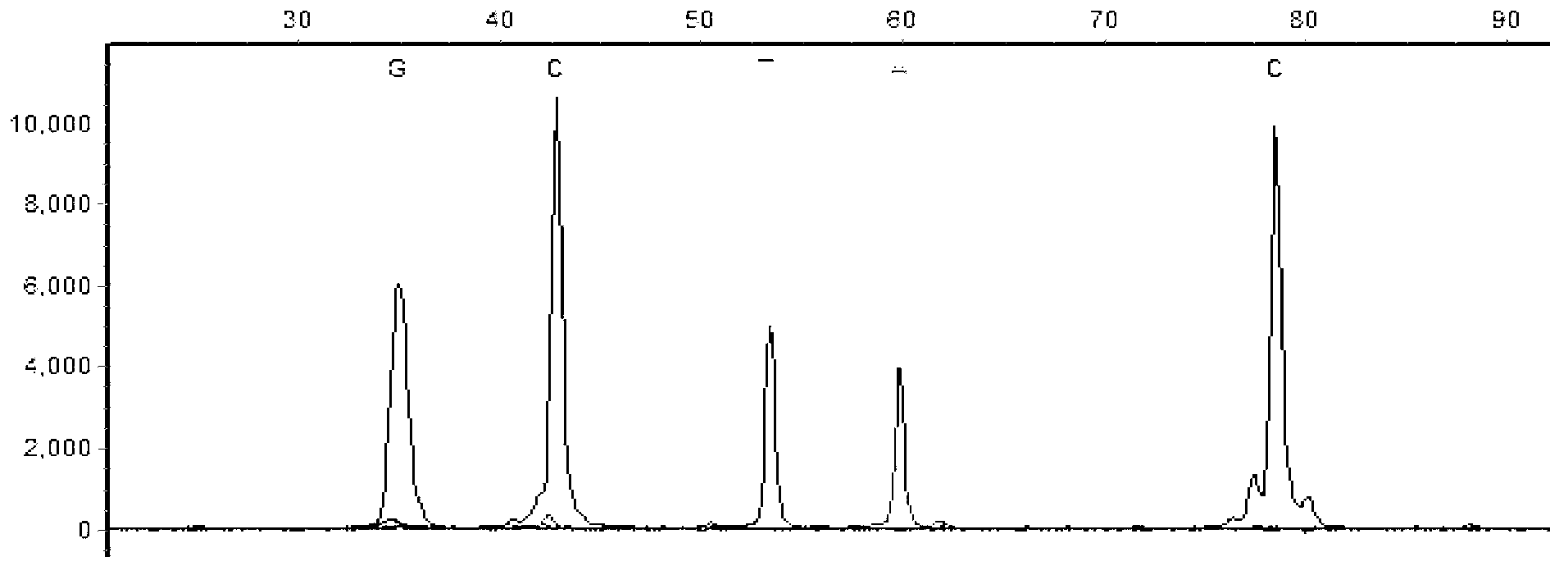
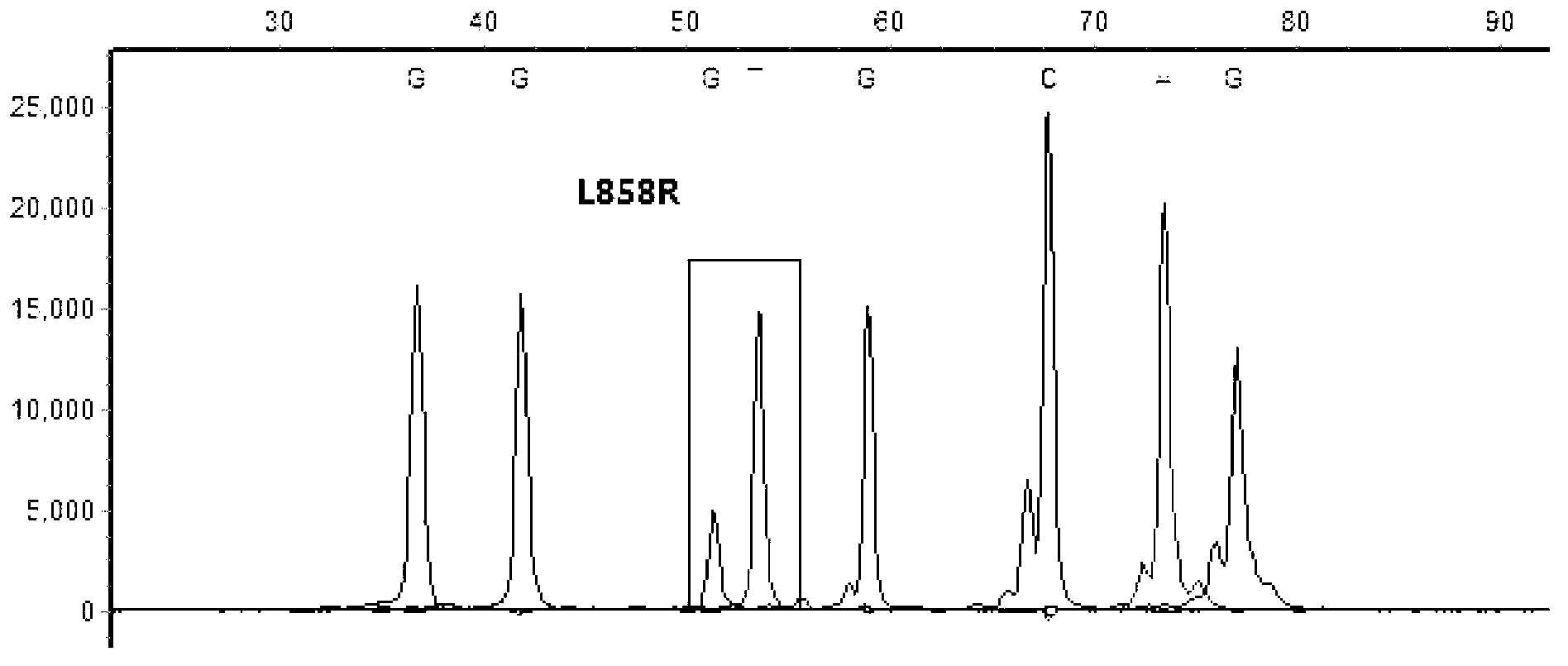
[0069] Primers were used to amplify the exon DNA segments containing hotspot mutation sites in seven driver genes of non-small cell lung cancer, BRAF, EGFR, PIK3CA, PTEN, MEK1, KRAS and NRAS, among which, the exon segment of BRAF BRAF_EX11, BRAF_EX15; the exon segment of EGFR is EGFR_EX18, EGFR_EX19, EGFR_EX20, EGFR_EX21; the exon segment of KRAS is KRAS_EX2; the exon segment of NRAS is NRAS_EX2, NRAS_EX3; the exon segment of PIK3CA PIK3CA_EX9, PIK3CA_EX20; the exon segment of PTEN is PTEN_EX5, PTEN_EX6, PTEN_EX7; the exon segment of MEK1 is MEK1_EX2.
[0070] The designed amplification primers for amplifying the exon segments of the seven driving genes of non-small cell lung cancer are shown in Table 6:
[0071] Table 6 Amplification primers (5'-3') of exon segments of related genes
[0072]
[0073] (2) Design extension primers: de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com