Corn photoperiod gene specific molecular marker and application thereof
A molecular labeling and photoperiod technology, applied in biochemical equipment and methods, DNA/RNA fragments, microbe determination/inspection, etc., can solve problems such as unconfirmed relationship with photoperiod and reduce environmental interaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] 1. Gene sequence:
[0048] Firstly, using the genomic DNA sequences of the ZmPIF3 genes of the corn varieties CML166 and Zheng 58 cloned by the present inventors, the nucleic acid sequences were compared by software Mega4.0.
[0049] 2. Genomic DNA extraction
[0050] Genomic DNA of maize inbred lines CML166 and Zheng 58, and genomic DNA of F2 populations crossed between CML166 and Zheng 58 were extracted by CTAB method.
[0051] 3. Design primers based on the differences in known related sequences:
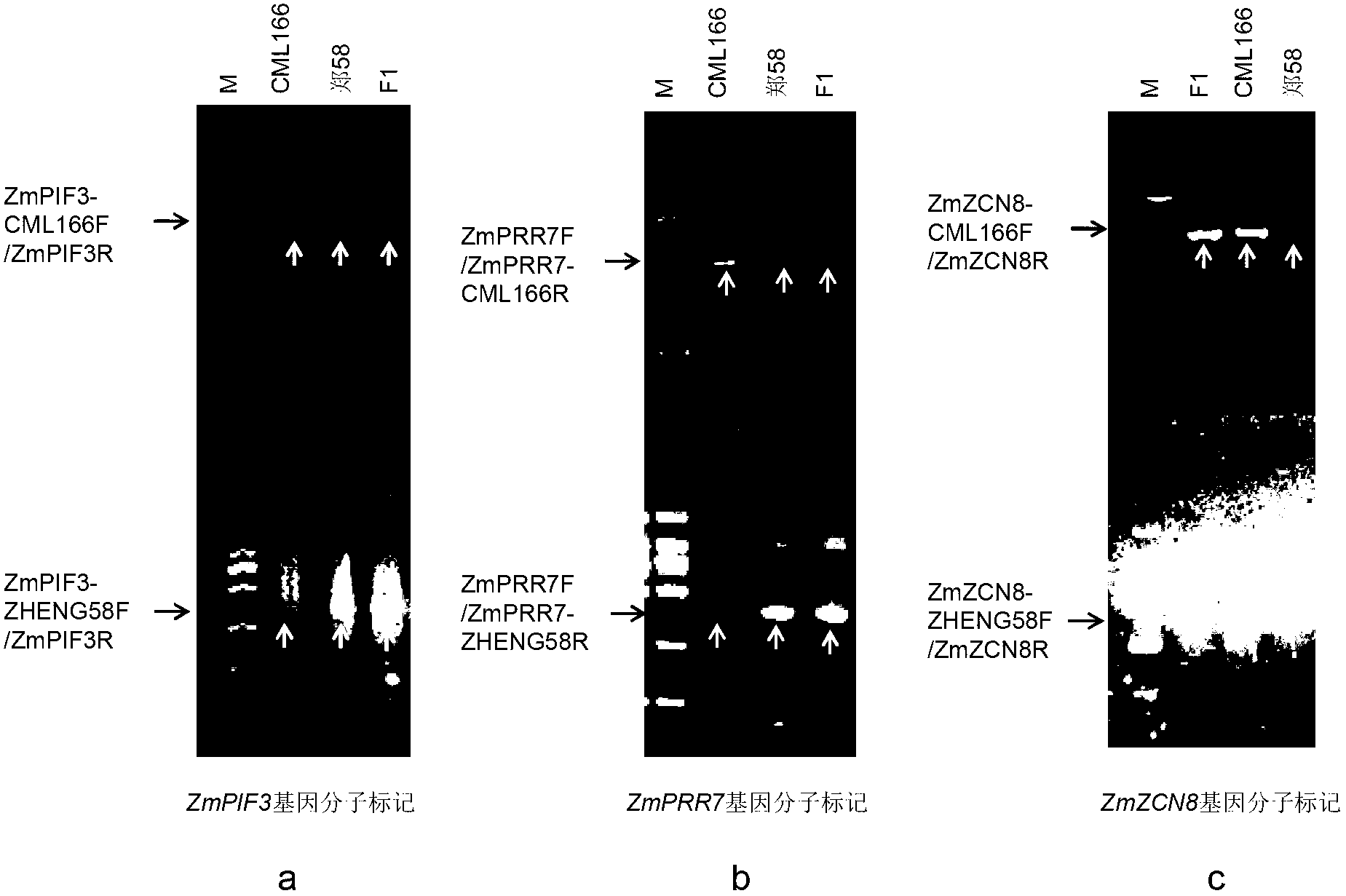
[0052] Comparing the partial DNA sequences of CML166 and Zheng 58 of the ZmPIF3 gene, the DNA sequence of the ZmPIF3 gene of CML166 is compared with the sequence of Zheng 58, and one of them has a transversion: the base of CML166 is G, while the base of Zheng 58 is a. Use the characteristics of sequence base transversion to design primers:
[0053] (1) ZmPIF3-CML166: forward: 5'-ACGCTGAGGGATGGATTGCTTG-3'
[0054] Reverse: 5'-AATGCACTGACAAGATGGTGGCTAT...
Embodiment 2
[0069] 1. Gene sequence:
[0070] First, use the genomic DNA sequences of the ZmPRR7 genes of the maize varieties CML166 and Zheng 58 obtained by cloning in the present invention, and use the software Mega4.0 to compare the nucleic acid sequences to obtain a sequence alignment.
[0071] 2, with the second step of embodiment 1.
[0072] 3. Design primers based on the differences in known related sequences:
[0073] Comparing the partial DNA sequences of CML166 and Zheng 58 in the ZmPRR7 gene by the software Mega4.0, the DNA sequence of the ZmPRR7 gene in CML166 is compared with the sequence of Zheng 58, and one of them has a transversion: the base of CML166 is T, and the base of Zheng The base of 58 is C (attached figure 2 ). Use the characteristics of sequence base transversion to design primers:
[0074] (1) ZmPRR7-CML166: forward: 5'-TTGAGAACCTTGTTTCACTAAAGGAT-3'
[0075] Reverse: 5'-CCGTAAGTGCTGCTATGAAGGTAT-3
[0076] (2) ZmPRR7-ZHENG58: forward: 5'-T...
Embodiment 3
[0084] 1. Gene sequence:
[0085] First, use the genomic DNA sequences of the ZmZCN8 genes of the maize varieties CML166 and Zheng 58 obtained by cloning in the present invention, and use the software Mega4.0 to compare the nucleic acid sequences to obtain a sequence alignment.
[0086] 2, with the second step of embodiment 1.
[0087] 3. Design primers based on the differences in known related sequences:
[0088] Comparing the partial DNA sequences of CML166 and Zheng 58 of the ZmZCN8 gene with the software Mega4.0, the DNA sequence of the ZmZCN8 gene of CML166 is compared with the sequence of Zheng 58, and one of them has a transversion: the base of CML166 is A, while the DNA sequence of Zheng 58 The base of 58 is G. Use the characteristics of sequence base transversion to design primers:
[0089] (1) ZmZCN8-CML166: Forward: 5'-GAGAGTTCTAATAAGAGCAACGA-3
[0090] Reverse: 5'-TGCAAGATTTAGGCATTTTGTTC-3'
[0091] (2) ZmZCN8-ZHENG58: Forward: 5'-GAGAGTTCTAA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com