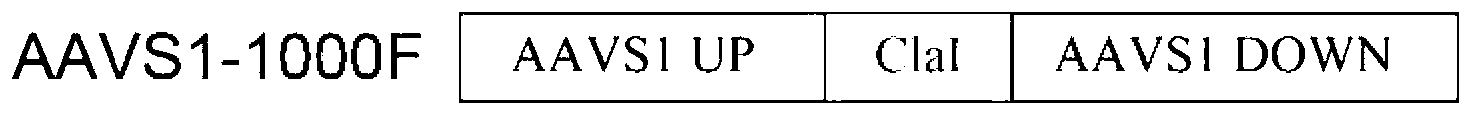Gene targeting system
A technology of gene targeting and targeting vector, applied in the biological field, can solve the problems of limited application and low efficiency, and achieve the effect of improving efficiency and level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Taking the gene targeting system targeting the AAVS1 site as an example, its composition is as follows:
[0022] 1. Zinc finger nuclease expression vector expressing AAVS1ZFN
[0023] The eukaryotic promoter CMV, the zinc finger nuclease AAVS1ZFN targeting the AAVS1 site, and the transcription termination signal TKpA were sequentially introduced into the multiple cloning site of the plasmid vector pshuttle (purchased from Agilent), wherein AAVS1ZFN contains the left zinc finger nuclease Two parts of AAVS1ZFNL and the right zinc finger nuclease AAVS1ZFNR, the left zinc finger nuclease AAVS1ZFNL and the right zinc finger nuclease AAVS1ZFNR are connected by self-cleaving polypeptide T2A (ie AAVS1ZFNL-T2A-AAVS1ZFNR), expressing the zinc finger of AAVS1ZFN For the structure of the nuclease expression vector see figure 1 . The zinc finger nuclease AAVS1ZFN targeting the AAVS1 site of this embodiment can also be used in the transcription activator-like effector nuclease (AAV...
Embodiment 2
[0076] Taking the gene targeting system targeting the CCR5 locus as an example, its composition is as follows:
[0077] (1) Zinc finger nuclease expression vector expressing CCR5ZFN
[0078] The zinc finger nuclease expression vector expressing CCR5ZFN consists of two vectors, one of which carries the left zinc finger nuclease CCR5ZFNL, and the other carries the right zinc finger nuclease CCR5ZFNR. The vector is based on the plasmid vector pshuttle, and the eukaryotic promoter CMV, the left zinc finger nuclease CCR5ZFNL or the right zinc finger nuclease CCR5ZFNR targeting the CCR5 site, and the transcription termination signal are sequentially introduced into the multiple cloning site of pshuttle TKpA constitutes a zinc finger nuclease expression vector expressing CCR5ZFN. The structure of the zinc finger nuclease expression vector is shown in Figure 5 .
[0079] See Sequence Listing 24 to gaaatcaact tctag 1035 for the sequence of CCR5ZFNL.
[0080] See sequence listing 2...
Embodiment 3
[0099]Taking the gene targeting system targeting the AAVS1 site as an example, its composition is as follows:
[0100] (2) The targeting vector targeting the AAVS1 site in this example consists of the following structure,
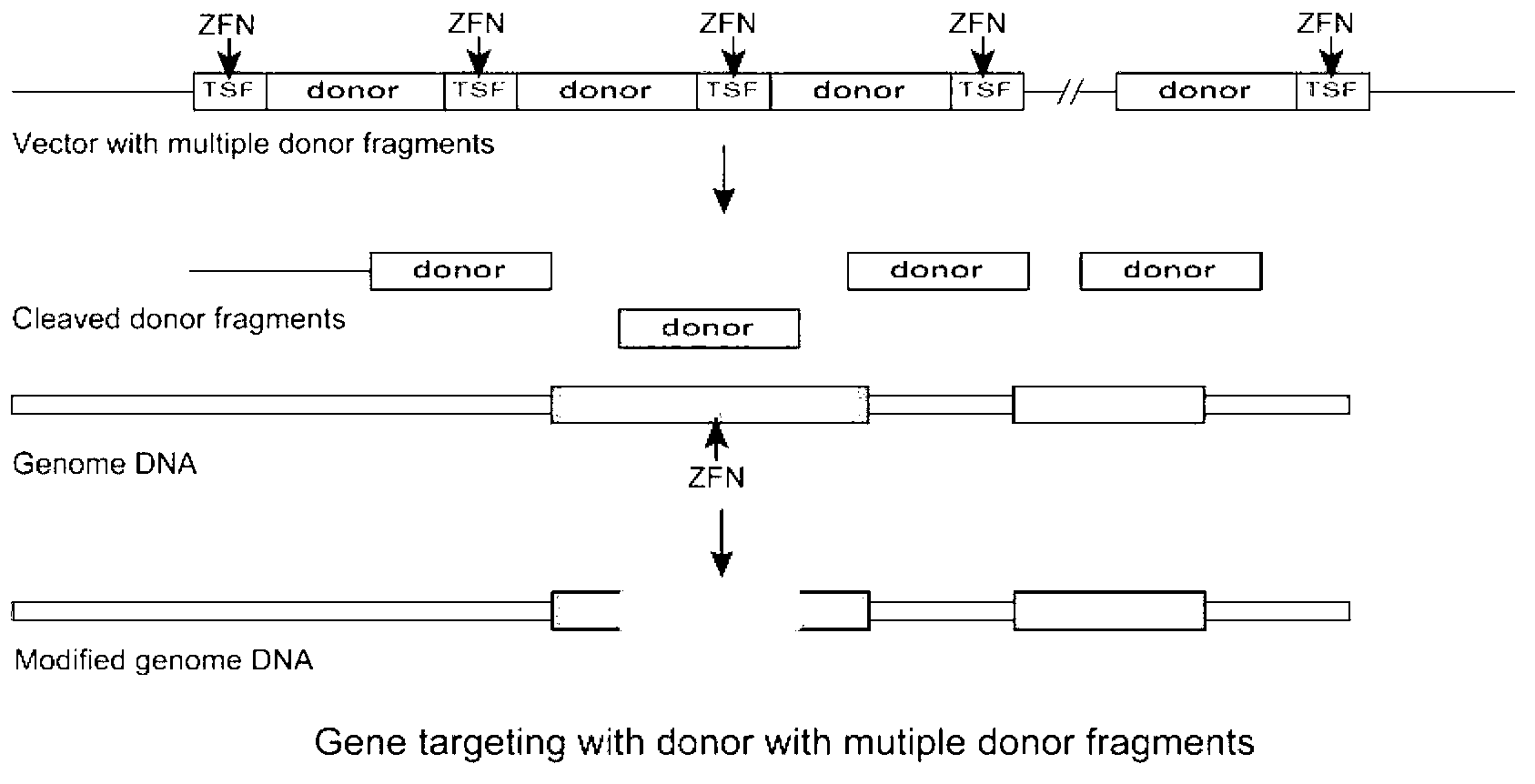
[0101] The multiple cloning site of pshuttle carries two donor DNA fragments AAVS1-2000F with tandem repeats, and the 5' and 3' ends of each donor DNA fragment are respectively inserted with the recognition sequence AAVS1-TSF of AAVS1ZFN, and the donor DNA The fragment consists of the upstream homology arm AAVS1UP, the downstream homology arm AAVS1DOWN and the exogenous DNA sequence (CMV-eGFP-T2A-luciferase-SV40pA, 3000bp fragment) between them. The upstream homology arm is the genome distance from AAVS1- TSF3000bp is a DNA sequence with a length of 1000bp, and the downstream homology arm is a DNA sequence with a length of 1000bp that is 100bp away from AAVS1-TSF on the genome.
[0102] See Sequence Listing 33 to gctgggttgg agtgg 5385 for the sequence of A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com