Expression and purification method for bombyx mori odorant binding protein (BmOBP2)
A technology for expression and purification of odorant binding protein, which is applied in the field of expression and purification of silkworm odorant binding protein BmOBP2, which can solve the problems of strong hydrophobicity, low abundance, and microscopic inhomogeneity, and achieve the effect of solving low expression level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Primer Design
[0050] Amplify the BmOBP2 target gene, the gene sequence is shown in SEQ ID NO.1, and the primers are designed as follows:
[0051] Upstream primer F: 5’- CGC GGATCC ATGAAGAGCAAAACAAAAC-3' (underlined restriction endonuclease Bam H I restriction site), as shown in SEQ ID NO.3;
[0052] Downstream primer R: 5'- CCG CTCGAG TTATAGTTCATCTTTAAC-3' (underlined restriction endonuclease xho I restriction site), as shown in SEQ ID NO.4;
Embodiment 2
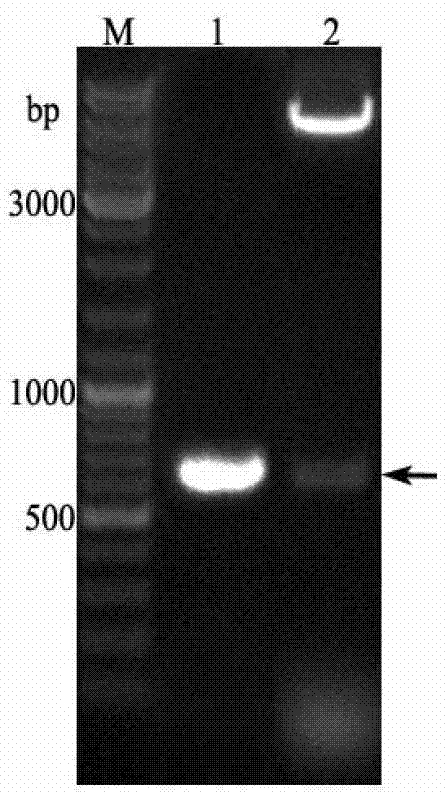
[0053] Example 2: Construction of pET-32a-BmOBP2 recombinant expression vector
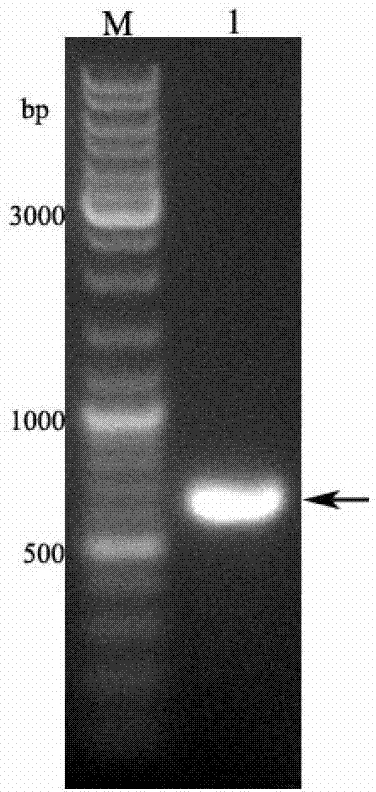
[0054] (1) Using the silkworm chrysalis cDNA as a template, use the primers F and R designed in Example 1 to amplify the specific fragment. The specific procedures are: pre-denaturation at 94°C for 5 minutes, denaturation at 94°C for 1 minute, annealing at 62°C for 45 seconds, and extension at 72°C 30s, cycled 30 times; finally extended at 72°C for 10 min, recovered product to obtain BmOBP2 gene (see figure 1 ), its nucleotide sequence is shown in SEQ ID NO.1.
[0055] In a 50 μL centrifuge tube, add the following components:
[0056] wxya 2 O 32.5 μL
[0057] TAQ Buffer 5μL
[0058] 2mM dNTPs 5μL
[0059] F 1.5μL
[0060] R 1.5 μL
[0061] cDNA 3μL
[0062] TAQ Polymerase (2U / μL) 1.5μL
[0063] After mixing the components evenly, put them into the PCR machine, and design 30 cycles according to the above reaction parameters. After the reaction is over, identify the amplified fragment by...
Embodiment 3
[0065] Embodiment 3: Expression of recombinant protein
[0066] The expression vector recombined in Example 2 was transformed into E.coli BL21 (purchased from Fermentas Company) to obtain the recombinant bacteria, in LB medium containing 50 mg / mL ampicillin (Shanghai Hengyuan Biotechnology Co., Ltd.) (purchased from Oxoid company) at 37°C, shaking at 220 rpm until OD 600 ≈0.5, add isopropyl-B-D-thiogalactopyranoside (IPTG) (final concentration 1 mM), induce culture at 37°C for 5 hours, take 1.5mL of bacterial liquid and centrifuge at 12000rpm, add 50μL of 20mM trimethylol Resuspend in aminomethane (Tris) (pH 8.0), then take 40 μL, add 10 μL 5× sample buffer, cook at 100°C for 10 minutes, and then centrifuge at 12000 rpm for 5 minutes to obtain recombinant protein samples, take 15 μL for SDS-PAGE, the results Such as Figure 3A As shown, it shows that the recombinant protein containing the HIS tag is successfully expressed by the above method and the expression level is very ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com