DNA (deoxyribonucleic acid) molecule, recombinant plasmid and escherichia coli
A DNA molecule, Escherichia coli technology, applied in the biological field, can solve the problems of inability to tolerate L-threonine, low protein content, and low acid production capacity of Escherichia coli
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Optimization and synthesis of DNA molecule of the present invention
[0032] The present invention obtains a protein sequence from Yersinia enterocolitica subsp.enterocolitica8081 from the NCBI database, its amino acid sequence is shown in SEQ ID NO: 4, and the NCBI protein number is Gi: 123440599. The present invention optimizes codons according to the codon preference of Escherichia coli W3110, synthesizes a DNA molecule with a nucleotide sequence as shown in SEQ ID NO: 1 through a nucleic acid synthesizer, verifies the correctness of the sequence by sequencing, and synthesizes a gene enzyme cut site Points are SmaI / HindIII. Using SmaI / HindIII as the insertion site, it was cloned into the high-copy plasmid vector pUC57, and Escherichia coli DH5α was used as the recipient strain for amplification.
[0033] The optimization and synthesis work were completed by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
Embodiment 2
[0034] Example 2: Construction of recombinant plasmid pKR-E
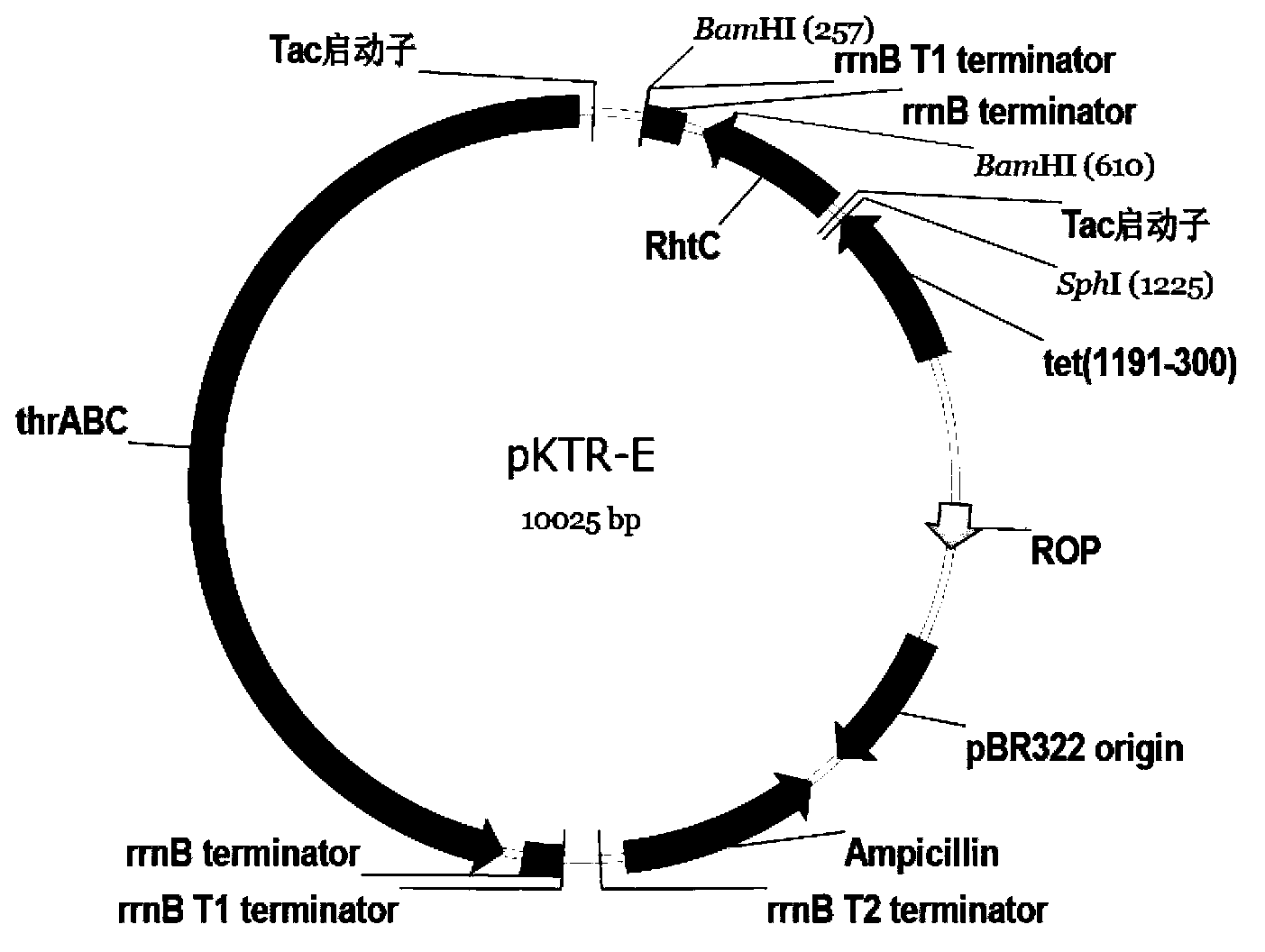
[0035] Extract the pUC57 plasmid from the above-mentioned Escherichia coli DH5α, digest with SmaI / HindIII, 50 μl system, T buffer+BSA, perform 1% agarose gel electrophoresis on the digested product, take off the gel at a suitable time, and cut the gel to recover the target gene fragment Take 3 μl and mix it with 1 μl of pKK223-3 plasmid that has been cut with SmaI / HindIII, and add 4 μl of high-efficiency ligase to make 8 μl of ligation system. Ligate overnight at 16°C, transform the prepared Escherichia coli Trans T1 competent cells the next day, screen with ampicillin at a final concentration of 100 μg / ml on an LB plate, pick a single colony after 20 hours of cultivation, and verify the plasmid by double digestion with HindIII and BamHI. See image 3 . The results show that the DNA molecule has been constructed to pKK223-3 to form a new recombinant plasmid pKR-E, see the plasmid map figure 1 .
[0036] Depend o...
Embodiment 3
[0037] Example 3: Construction of recombinant plasmid pKTR-E
[0038] Extract the plasmid pKR-E in Escherichia coli in Example 2 as a template, and use the primers shown in SEQ ID NO: 2 and SEQ ID NO: 3 in the sequence listing to amplify the target gene fragment, which has a section on pKTR-E Tac promoter sequence, rrnB T1 Terminator, rrnB Terminator terminator sequence derived from basic plasmid pKK223-3. BamHI / SphI digested amplified product, took 3 μl and mixed it with 1 μl of pKK223-3-thrA’BC plasmid that had also been double-digested with BamHI / SphI, and added 4 μl of high-efficiency ligase to make 8 μl of ligation system. Ligate overnight at 16°C, transform the prepared Escherichia coli Trans T1 competent cells the next day, screen with ampicillin at a final concentration of 100 μg / ml on an LB plate, pick a single colony after 20 hours of cultivation, and extract the plasmid for HindIII digestion verification, see image 3 . The results show that the fragment has been ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com