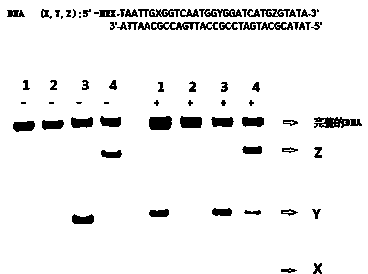
Method for detecting 5-aldehyde cytosine and 5-hydroxymethylcytosine in DNA (deoxyribonucleic acid) by utilizing piperidine aqueous solution
A technology for hydroxymethylcytosine and aldehydecytosine, applied in the field of detection of 5-formylcytosine and 5-hydroxymethylcytosine in DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
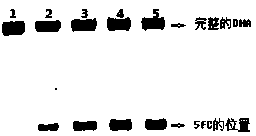
[0016] Example 1: Reaction of 5-formylcytosine DNA with 5% piperidine
[0017] Take 2 μL of 5-formylcytosine DNA (10 μM), add 93 μL of water and 5 μL of piperidine, heat-treat at 90°C for 0.5 hours, spin the solvent, load the sample to the electrophoresis tank, and observe the gel results after 2 hours , 10% of the 5-formylcytosine DNA was fragmented.
Embodiment 2
[0018] Example 2: Reaction of 5-formylcytosine DNA with 10% piperidine
[0019] Take 2 μL of 5-formylcytosine DNA (10 μM), add 88 μL of water and 10 μL of piperidine, heat-treat at 90°C for 0.5 hours, spin the solvent, load the sample to the electrophoresis tank, and observe the gel results after 2 hours , 35% of the 5-formyl cytosine DNA fragmentation occurred.
Embodiment 3
[0020] Example 3: Reaction of 5-formylcytosine DNA with 15% piperidine
[0021] Take 2 μL of 5-formylcytosine DNA (10 μM), add 83 μL of water and 15 μL of piperidine, heat-treat at 90°C for 0.5 hours, spin the solvent, load the sample to the electrophoresis tank, and observe the gel results after 2 hours , 50% of the 5-formylcytosine DNA fragmentation occurred.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com