Novel affinity ligand polypeptide library of immunoglobulin G constructed based on protein A affinity model and application of design method
A technology of immunoglobulin and design method, which is applied in the field of computer simulation and protein separation and purification, and can solve the problems of poor specificity and affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Obtaining the Simplified Affinity Binding Model of SpA
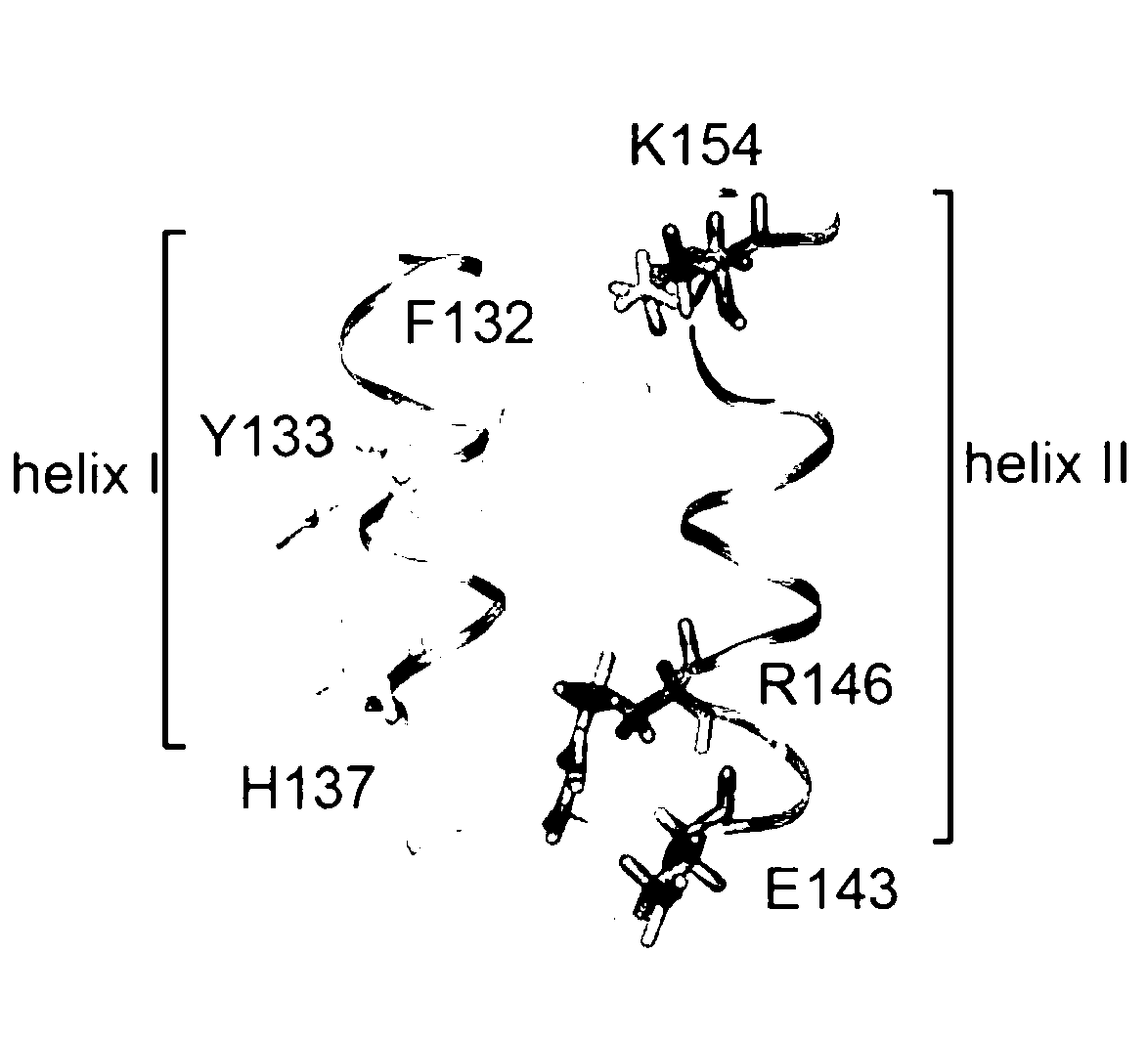
[0050] The absolute binding free energy of the S. aureus protein SpA and hIgG1 complex was first calculated using the MM / PBSA method. Then, a free energy decomposition method based on MM / PBSA was used to analyze the molecular mechanism of the high affinity between SpA and hIgG1 and to analyze the contribution of the residues on the binding surface of the SpA-hIgG1 complex to the binding free energy. Hotspot residues for the interaction of SpA and hIgG1 were identified based on the free energy contribution of each residue and the analysis of residue interaction pairs. Finally, an affinity binding model of SpA was constructed based on the molecular mechanism and hotspot residues obtained from the above analysis.
[0051] The SpA-hIgG1 complex molecular system was used for molecular dynamics simulations. Each complex model contains a single chain of the Fc fragment and the B domain of SpA. The model stru...
Embodiment 2
[0066] The construction of embodiment 2 polypeptide library
[0067] 1. Determine the length of the peptide sequence
[0068] A known:
[0069]
[0070]
[0071]
[0072]
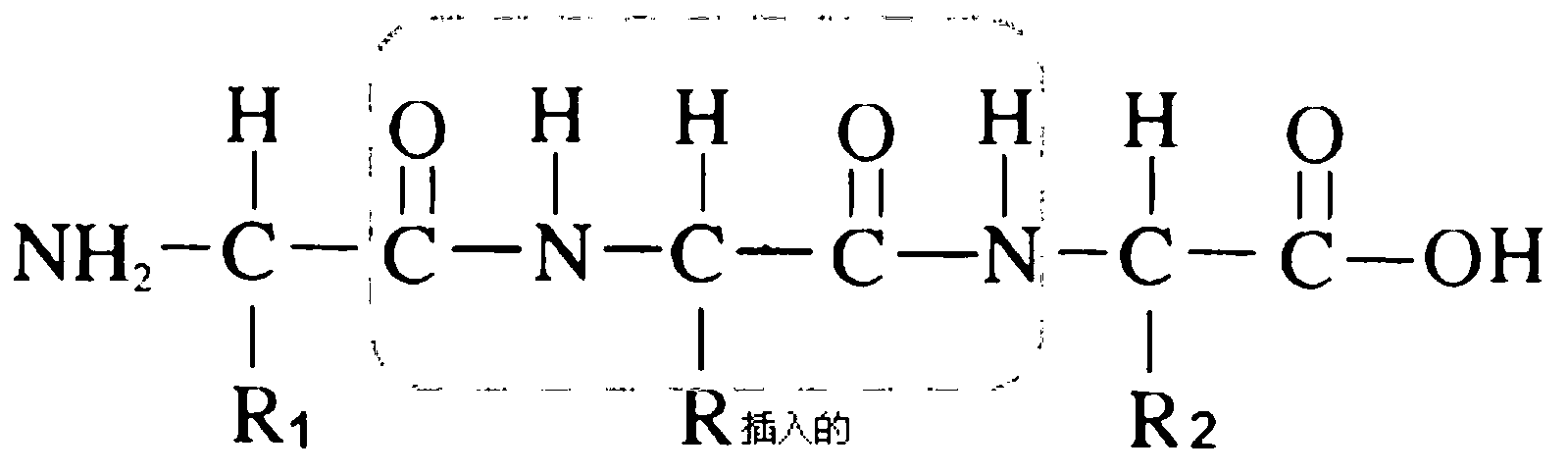
[0073] image 3 Example of insertion of one amino acid (the line inside the dotted box represents the added bond). If the distance between the C and N termini between two hotspot residues , an amino acid residue can be inserted; if Insertion of two amino acid residues is considered; if Three or more amino acid residues need to be inserted.
[0074] According to the distance between the corresponding N and C terminals of the two hot spot residues of SpA (calculated by using Visual Molecular Dynamics, VMD software), determine the number of amino acids to be inserted. The final peptide construction mode is shown in Table 1.
[0075] Table 2
[0076] Table 1
[0077]
[0078]Among them, one mode is an octapeptide, and the rest are heptapeptides. Considering that the N and C ends of...
Embodiment 3
[0086] Example 3 Docking of Polypeptide Molecule and Fc Fragment
[0087] 1. Vina docking of polypeptide and Fc fragment
[0088] Since a single residue has a strong affinity with Fc, it does not necessarily mean that the polypeptide composed of residues has a strong interaction with Fc, so continue to use vina to sequentially dock all the polypeptides in the polypeptide library with the CBC of the Fc fragment. Docking results show that all peptides can bind to Fc fragments, and the predicted binding free energy of all peptides is between -4.5~-8.2kcal / mol, which is in line with the moderate affinity requirements of affinity ligands (the binding constant is between 10 4 ~10 8 m -1 between). Among them, the peptides with binding free energy around -6.5kcal / mol were the most distributed. In order to avoid missed selection during screening, polypeptide molecules with binding free energy lower than -6.5kcal / mol were selected, and a total of 754 polypeptides were selected. For...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com