Method and reagent for adjusting methylation/demethylation state of nucleic acid
A technology of demethylation and glycosylase, applied in the fields of biotechnology and pharmacy, which can solve unknown problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0228] Preparation of Flag-MBD4: The method is similar to the method for preparing and purifying Flag-TDG, and the MBD4 protein sequence is shown in GenBank accession number AAC68878.
[0229] Preparation of Flag-UNG2: The method is as in the preparation and purification of Flag-TDG, and the UNG2 protein sequence is shown in GenBank accession number NP_001035781.
[0230] Preparation of GST-SUMG1: The coding sequence of SUMG1 was amplified by PCR, digested with BamHI and XhoI and inserted into the BamHI and XhoI sites of pGEX-4T vector. pGEX-4T-SMUG1 was transformed into BL21(DE3), and protein expression was induced with IPTG. GST-SMUG1 was purified with Gluthione Sepharose 4B according to the GE healthcare product manual; the SUMG1 protein sequence is shown in GenBank accession number AAL86910.
[0231] The method for purifying 6XHis-fused TDG protein from bacteria: subcloning the TDG coding sequence from the pcDNA4-Flag vector to the BamHI and XhoI sites of the pET28a vecto...
Embodiment 1
[0271] Example 1. The modification activity of 5mC and 5hmC was detected in the nuclear extract of mammalian cells transfected with Tet2
[0272] It has been reported that Tet dioxygenase can hydroxylate 5mC to 5hmC (M. Tahiliani et al., Science 324, 930 (May 15, 2009)), but it is not clear whether it is in nuclear extracts of mammalian cells Has modification activity against 5mC and / or 5hmC. Due to the uncertainty of the presence or absence of this modification activity, choosing an appropriate method is critical to the success of the assay.
[0273] with radioactive isotopes 32 P-terminal labeling of modified nucleotides digested with restriction endonucleases, followed by thin layer chromatography (thin layer chromatography, TLC) analysis, is a very sensitive method. However, base modifications may hinder endonuclease digestion. In order to solve this problem, the inventors have utilized a feature of the restriction endonuclease EcoNI, that is, it can recognize sequences...
Embodiment 2
[0276] Example 2, the modification of 5mC and 5hmC in DNA is catalyzed by Tet dioxygenase
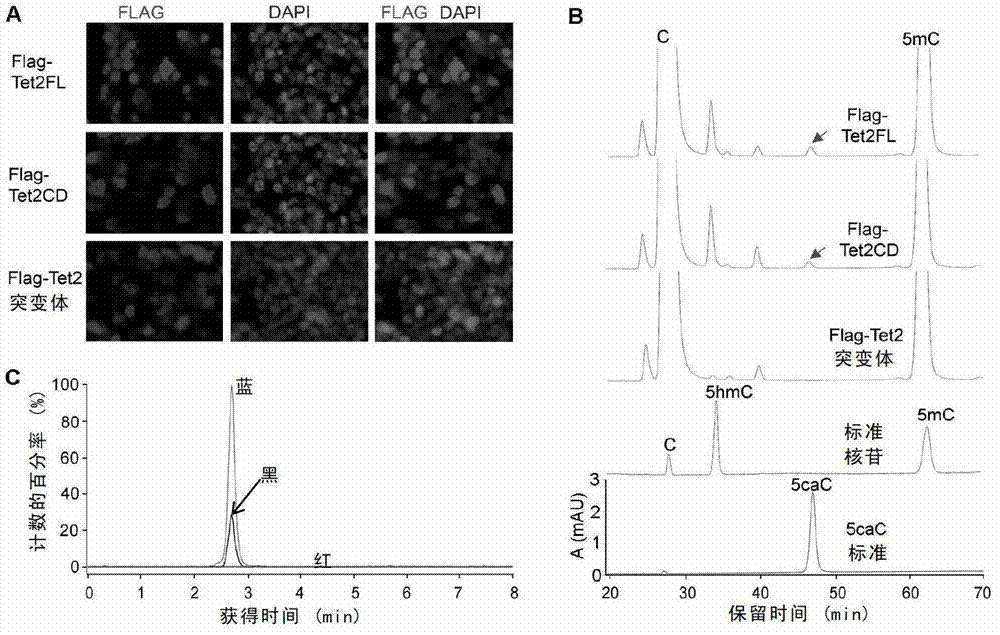
[0277] Since Tet dioxygenase can catalyze the oxidation of 5mC to 5hmC, the inventors speculated that the unidentified nucleotide "X" spot detected on the TLC plate might be catalyzed by a protein interacting with Tet2 or Tet2 itself. In order to test this conjecture, the inventors transfected the full-length Tet2 with Flag tag in 293T cells (i.e. transfected with the expression plasmid expressing Flag-Tet2), and purified Tet2 protein to detect whether it has DNA containing 5mC modification activity. TLC analysis showed that after the DNA substrate containing 5mC or 5hmC reacted with the purified Tet2 protein, it was detected that an "X" point ( figure 1 A).
[0278] In order to confirm that the "X" point on the TLC plate is derived from 5mC, the inventors performed an isotope tracer experiment. Using the CpG-specific bacterial methyltransferase M.SssI and [ 14 C methyl] S-adenosylm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com