Aptamer for identifying zeatin through specifity, and screening method and application of aptamer
A nucleic acid aptamer and zeatin technology, applied in biochemical equipment and methods, microbial measurement/testing, organic chemistry, etc., can solve problems such as small molecular weight
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
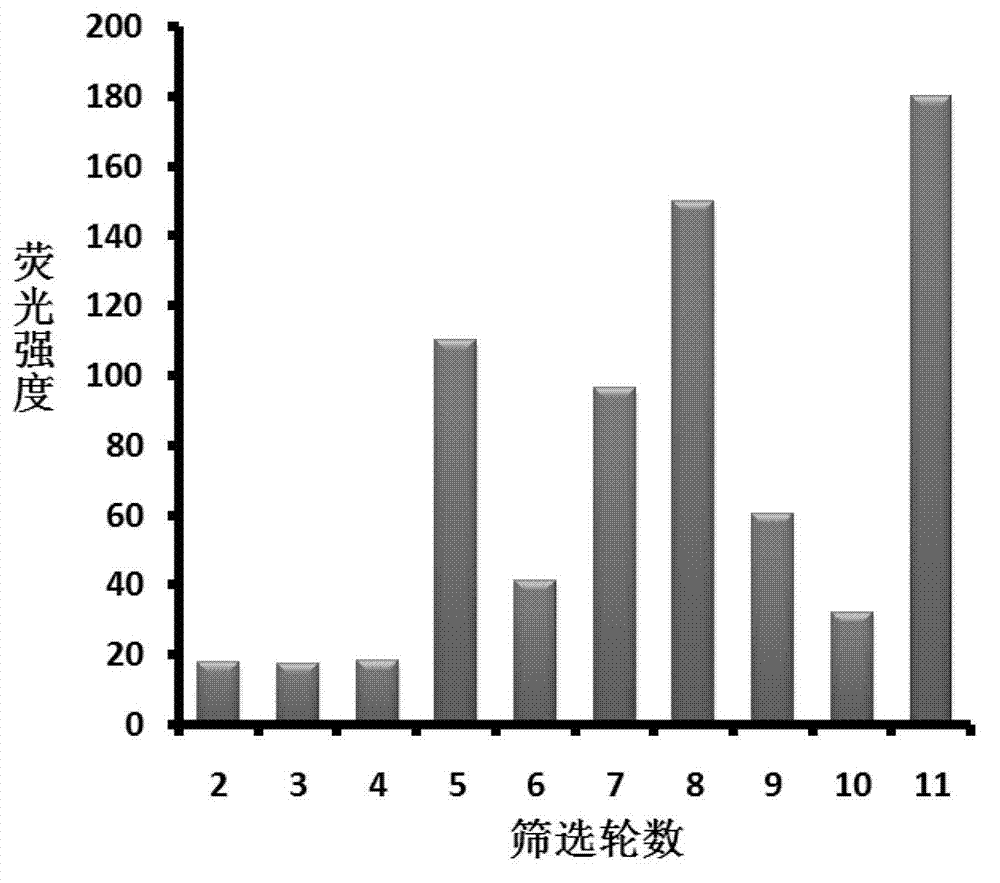
[0095] Embodiment 1, the screening of zeatin nucleic acid aptamer
[0096] 1. Fixation of L-histidine and zeatin
[0097] 0.5 g of epoxy-activated Sepharose 6B (GE Healthcare, Sweden) microspheres were repeatedly washed with 100 mL of water to obtain 1.75 mL of wet spheres, which were then washed with 0.2 M Na 2 CO 3 (pH≈12) to replace the water in the swollen microspheres, add 45mM L-histidine or amino-modified trans-zeatin (see below for the synthesis steps) to the 3.5mL reaction system, shake and react at room temperature for 48h . After the reaction, use pH4.5 sodium acetate buffer solution (0.1M sodium acetate, 0.5M NaCl) and pH12 sodium carbonate buffer solution (0.2M NaHCO 3 / Na 2 CO 3 , 0.5M NaCl; that is, use 0.2M NaCl 2 CO 3 Add HCl to the solution to adjust the pH to 12, and at the same time make the concentration of NaCl 0.5M) Repeatedly wash the microspheres alternately, finally wash with water, and dilute to 3.5mL, store in a refrigerator at 4°C.
[0098]...
Embodiment 2
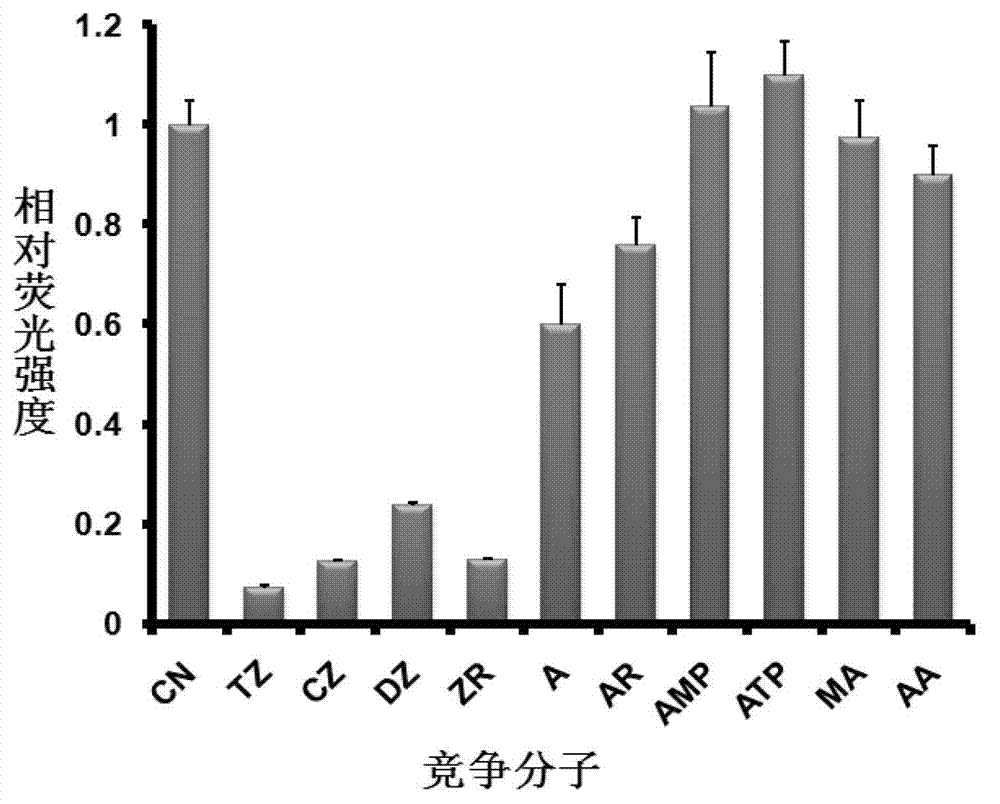
[0141]Embodiment 2, the specific detection of zeatin nucleic acid aptamer
[0142] 1. Nucleic acid aptamer pretreatment
[0143] 0.5 μM 6-carboxyfluorescein (6-FAM)-labeled aptamer 2 (sequence 2) was dissolved in binding buffer, denatured at 95°C for 5 minutes, cooled on ice for 15 minutes, and left at room temperature for 5 minutes.
[0144] 2. Nucleic acid aptamer specific detection
[0145] Mix 5 μL of zeatin microspheres (microspheres immobilized with trans-zeatin obtained in step 1 in Example 1) with 1 mM zeatin (cis-zeatin (CZ), trans-zeatin (TZ), dihydrogen Zeatin (DZ), trans-zeatin nucleoside (ZR)) and its analogues (adenine (A), adenosine (AR), adenosine monophosphate (AMP), adenosine triphosphate (ATP), ethanolamine ( AA), 2-methallyl alcohol (MA)) was added to the aptamer solution pretreated in step 1 (zeatin and its analogues were used as aptamer competitors), and incubated at room temperature 30min, heated at 95°C for 5min with the elution buffer, eluted the DN...
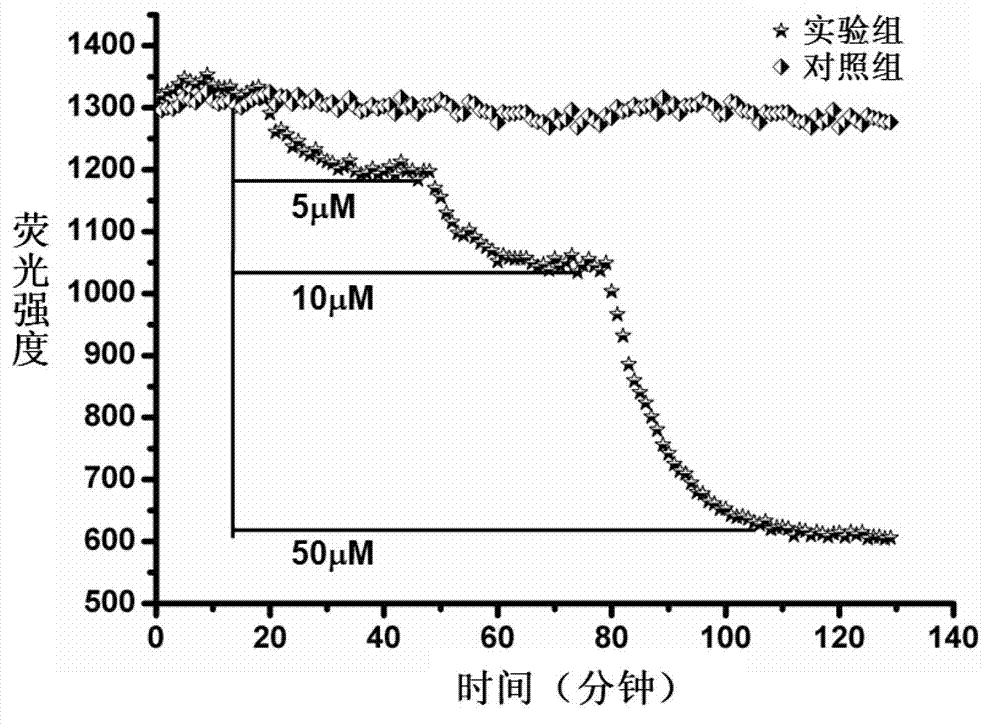
Embodiment 3
[0147] Embodiment 3, the preparation of nucleic acid probe
[0148] The following four sets of nucleic acid probes were synthesized by Shanghai Bioengineering Company:
[0149] Nucleic acid probe A: composed of two DNA fragments (DNA fragment A-1 and DNA fragment A-2); DNA fragment A-1 is nucleic acid aptamer 1 (sequence 1), and its 5' end has 6-FAM ( Fluorescent reporter group), BHQ1 (fluorescent quencher group) at the 3' end, referred to as the aptamer element; DNA fragment A-2 is a partial reverse complementary sequence of the nucleic acid aptamer 1 (sequence 9), referred to as the complementary element;
[0150] Nucleic acid probe B: composed of two DNA fragments (DNA fragment B-1 and DNA fragment B-2); DNA fragment B-1 is nucleic acid aptamer 1 (sequence 1), and its 5' end has 6-FAM ( Fluorescent reporter group), referred to as aptamer element; DNA fragment B-2 is a partial reverse complementary sequence of aptamer 1 (SEQ ID NO: 10), and its 3' end has BHQ1 (fluorescent ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com