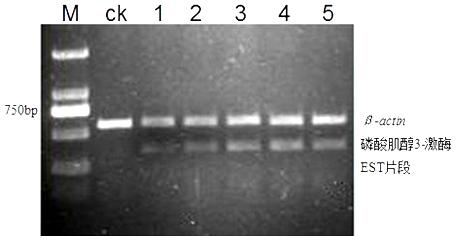
Phosphoinositide 3-kinase EST segment in common wild rice expressed by bacterial blight stress
A technology of bacterial blight bacteria and phosphoinositide, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, transferases, etc., can solve the problem of difficult acquisition of bacterial blight resistance genes and loss of bacterial blight resistance genes Resistance, no effective measures, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Obtaining the EST fragment of phosphoinositide 3-kinase expressed in common wild rice under the stress of bacterial blight
[0057] 1. Design primers to amplify the phosphoinositide 3-kinase EST fragment expressed in common wild rice under the stress of bacterial blight
[0058] According to the obtained phosphoinositide 3-kinase EST fragment information SEQ ID NO: 1, use the biological software Primer5.0 to design primers, which are composed of upstream primers and downstream primers. The base sequence of the upstream primers is as in the sequence listing As shown in SEQ ID NO: 2, the base sequence of the downstream primer is shown in SEQ ID NO: 3 in the sequence listing. The primers were synthesized by Huada Gene Company.
[0059] 2. Total RNA was extracted from common wild rice leaves inoculated with bacterial blight
[0060] (1) Processing of samples
[0061] After the common wild rice leaves were inoculated with the bacteria solution of bacterial bl...
Embodiment 2
[0093] Example 2 A method for detecting the differential expression of phosphoinositide 3-kinase EST fragments in common wild rice provided by the present invention comprises the following steps:
[0094] 1. Extraction of total RNA
[0095] 1) Processing of samples
[0096] A. Treatment of the samples to be tested: After inoculating common wild rice leaves with bacterial blight liquid, collect and inoculate the common wild rice leaves at 24 h, 48 h, 72 h, 96 h, and 120 h at intervals of 24 h. The leaves of wild rice were placed in a mortar and quickly ground with liquid nitrogen, and 1 ml of TRIzol extract was added to each 100 mg sample, and vortexed for 15 s. The sampling of each period was a treatment sample, a total of 5 treatment samples;
[0097] B. The treatment of the control group, using sterilized double-distilled water instead of the bacteria liquid of bacterial blight to inoculate the leaves of common wild rice as the control group, every 24 h, at 24 h, 48 h, 72 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com