Distributed gene sequence alignment method based on Basic Local Alignment Search Tool (BLAST)
A gene sequence, distributed technology, applied in the field of computer and bioinformatics, can solve the problems affecting the overall execution efficiency of software, high hardware cost, high network resource overhead, etc., to reduce the IO bottleneck effect, increase the cumulative IO bandwidth, reduce The effect of hardware cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The specific implementation manners of the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. The following examples are used to illustrate the present invention, but are not intended to limit the scope of the present invention.
[0028] Method flow chart of the present invention is as figure 1 shown, including the following steps:
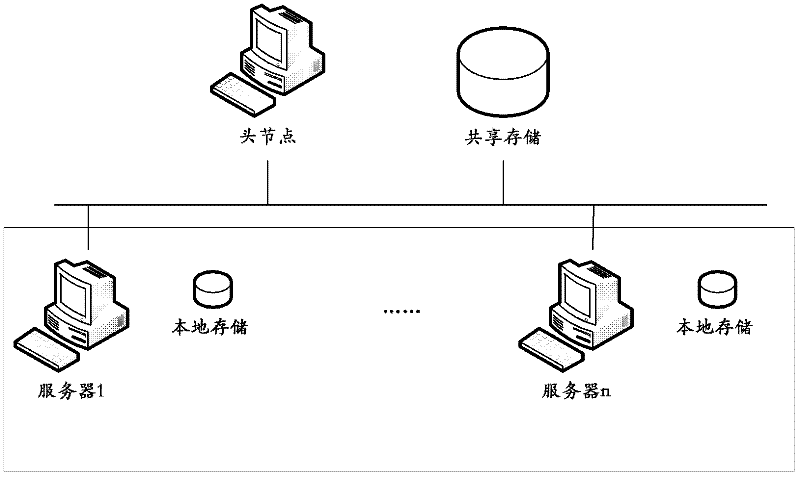
[0029] S1, first the program parses the user parameters, and determines the number of MPI (Message Passing Interface, message passing interface) threads, reads the query sequence file (FASTA format) and divides the query sequence file according to the number of tasks (that is, the query sequence file) ( The number of tasks is greater than or equal to the number of MPI threads) to obtain the query sequence file segment, and then each MPI thread reads its own MPI line program number respectively; the user parameters mainly refer to BLAST parameters, and BLAST is an open ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com