Method for concentrating sample constituents and amplifying nucleic acids
A nucleic acid and biological sample technology, applied in biochemical equipment and methods, analytical materials, recombinant DNA technology, etc., can solve problems such as difficulty in the preparation of μ-TAS, achieve time and cost savings, and reduce the number of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
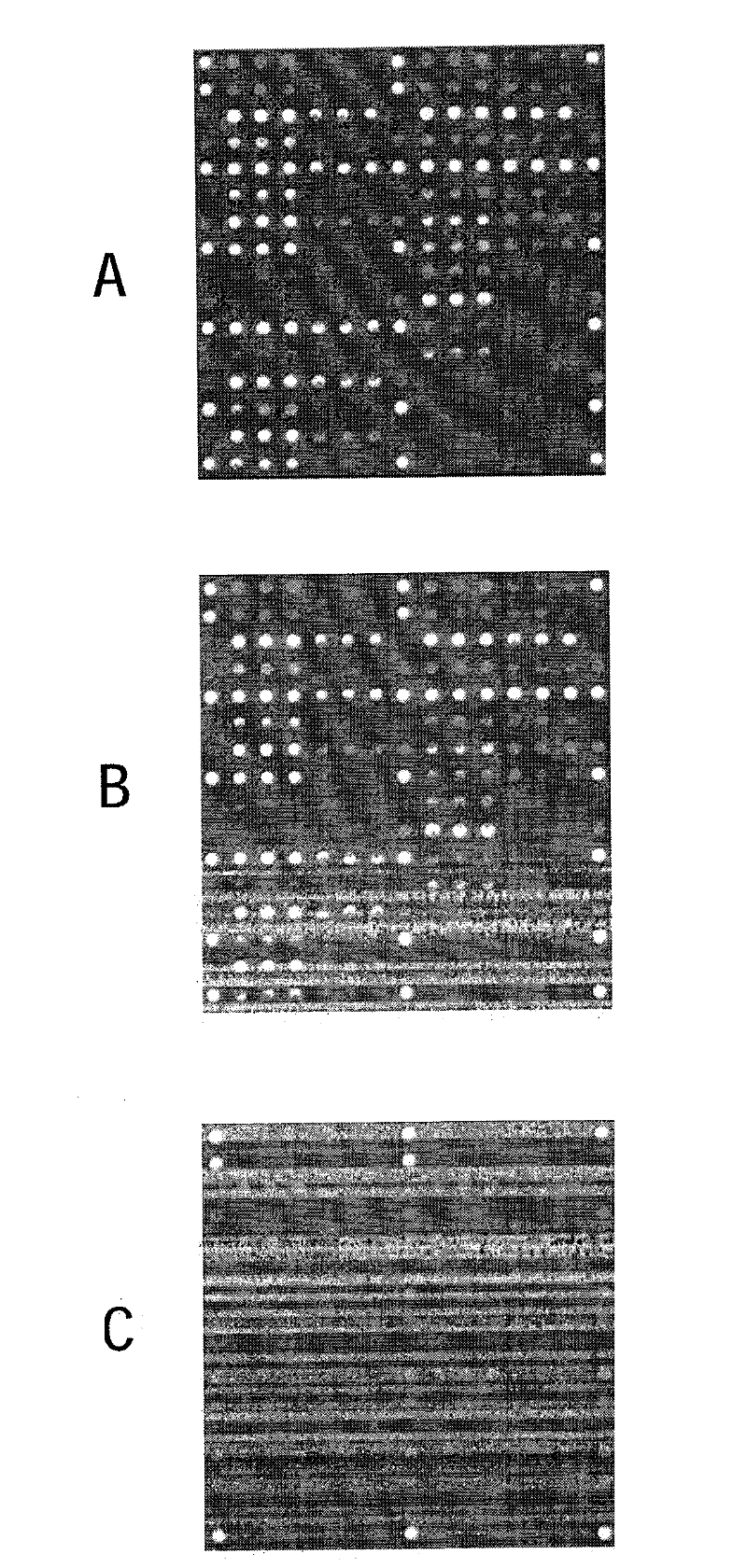
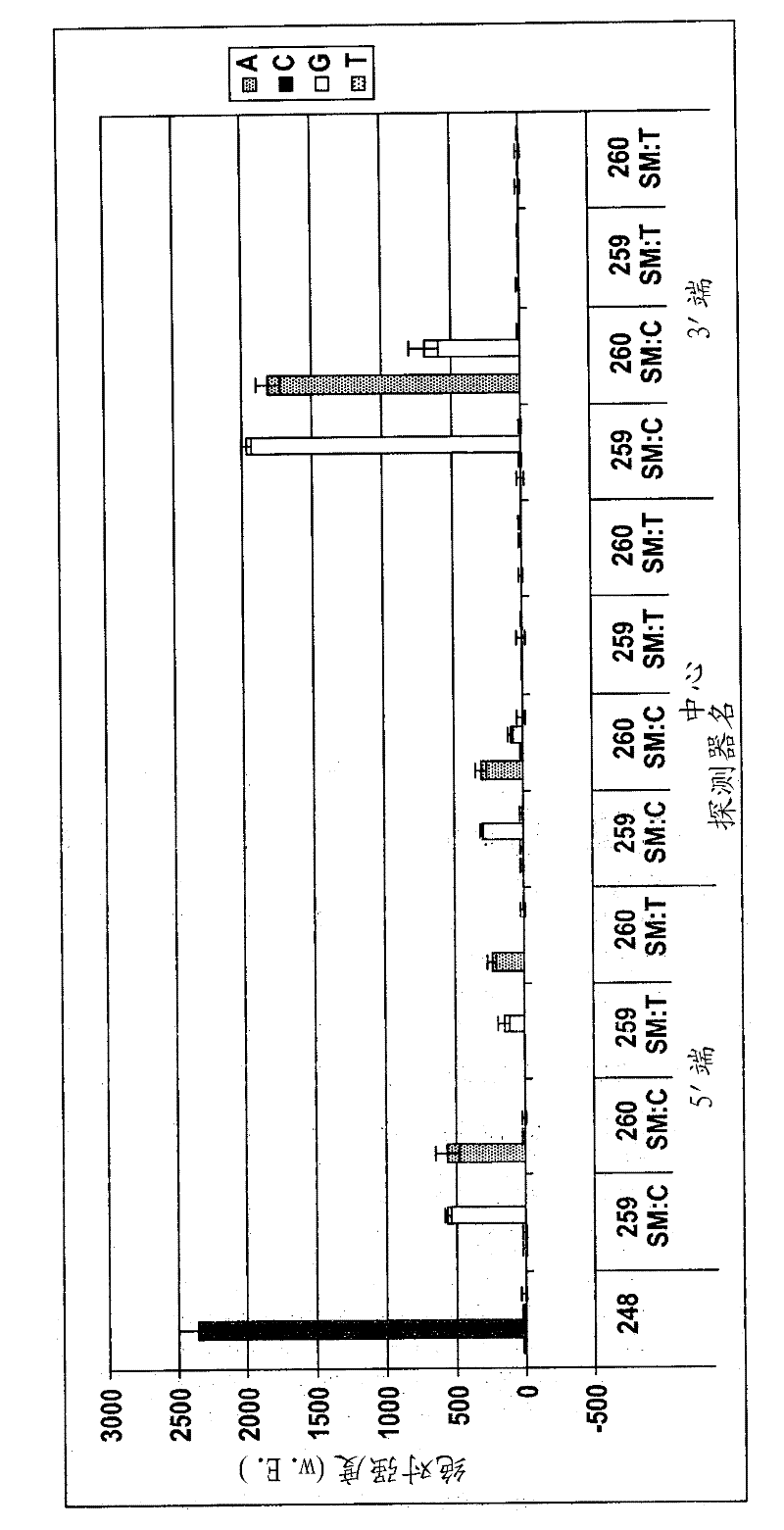
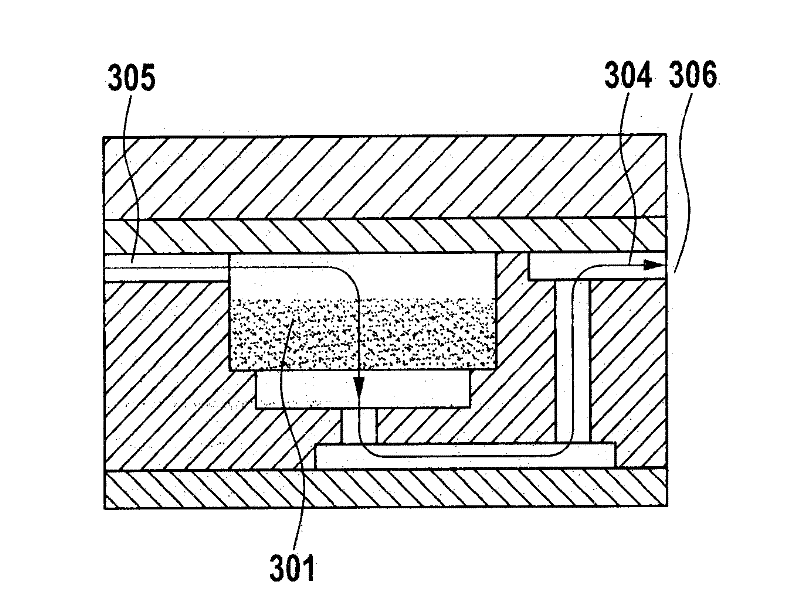
[0076] figure 1 A schematic flow diagram of the method of the invention using the separation system is shown.
[0077] A biological sample such as urine (which contains germs such as E. coli) is applied to the filter. The nucleic acid to be amplified is DNA, and the amplification is performed by means of PCR.
[0078] In step 1, each ml contains 10 4 -10 7 A macroscopic sample volume (10 ml) of the bacterial suspension 102 of 1 bacteria is passed through the filter (here the silica fiber mat 101 ) at a pressure of maximum 0.5 bar for 10 minutes (ie 1 ml / min). E. coli is retained on the silica fiber mat with a separation of 95%-99%, so that the suspension 103 then contains almost no bacteria 102. Then in step B, wash the filter 101 with 100-500 μl of PCR buffer to create optimal conditions for PCR. In step C, the PCR solution, ie 30 μl PCR master mix containing dNTPs, buffer, Taq-polymerase, primers and PEG, is added and the PCR cycle is started. In the PCR reaction, the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
| separation | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com