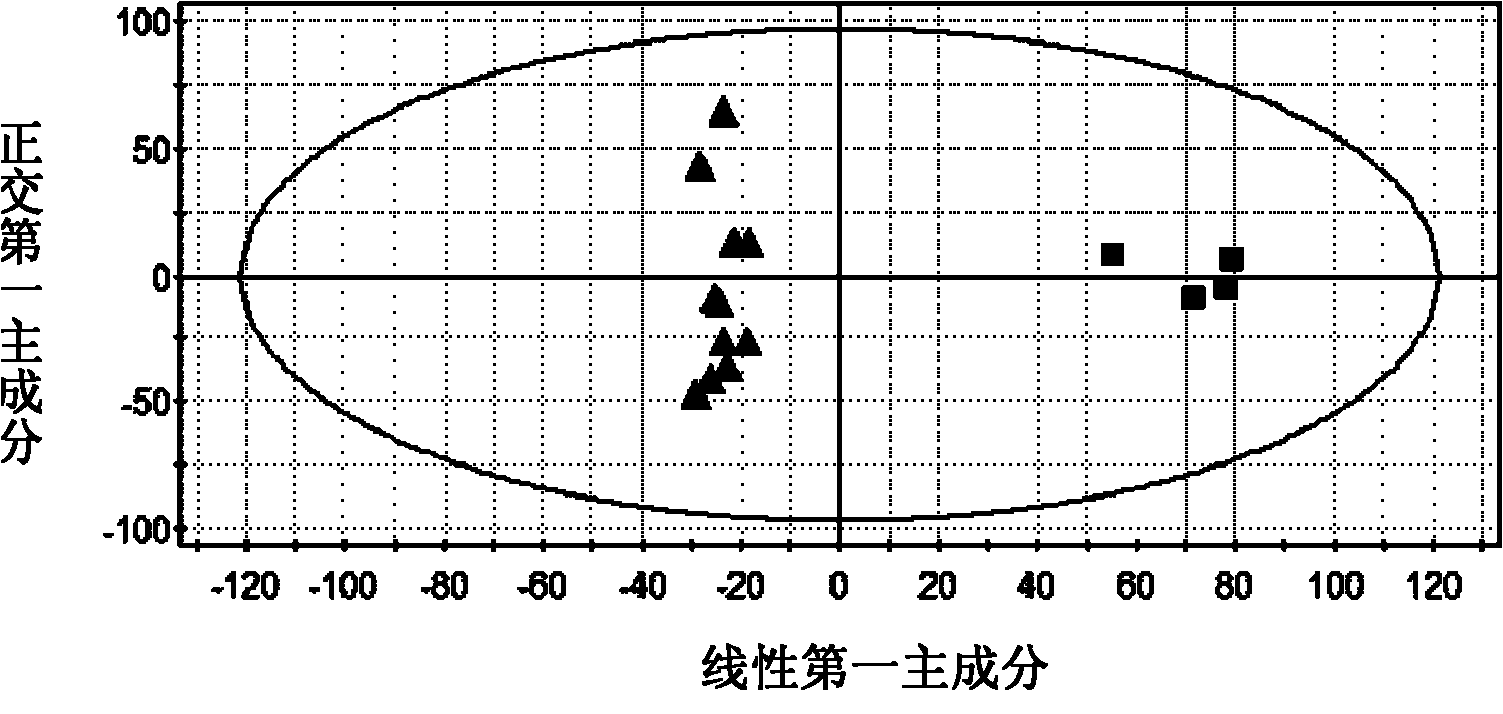
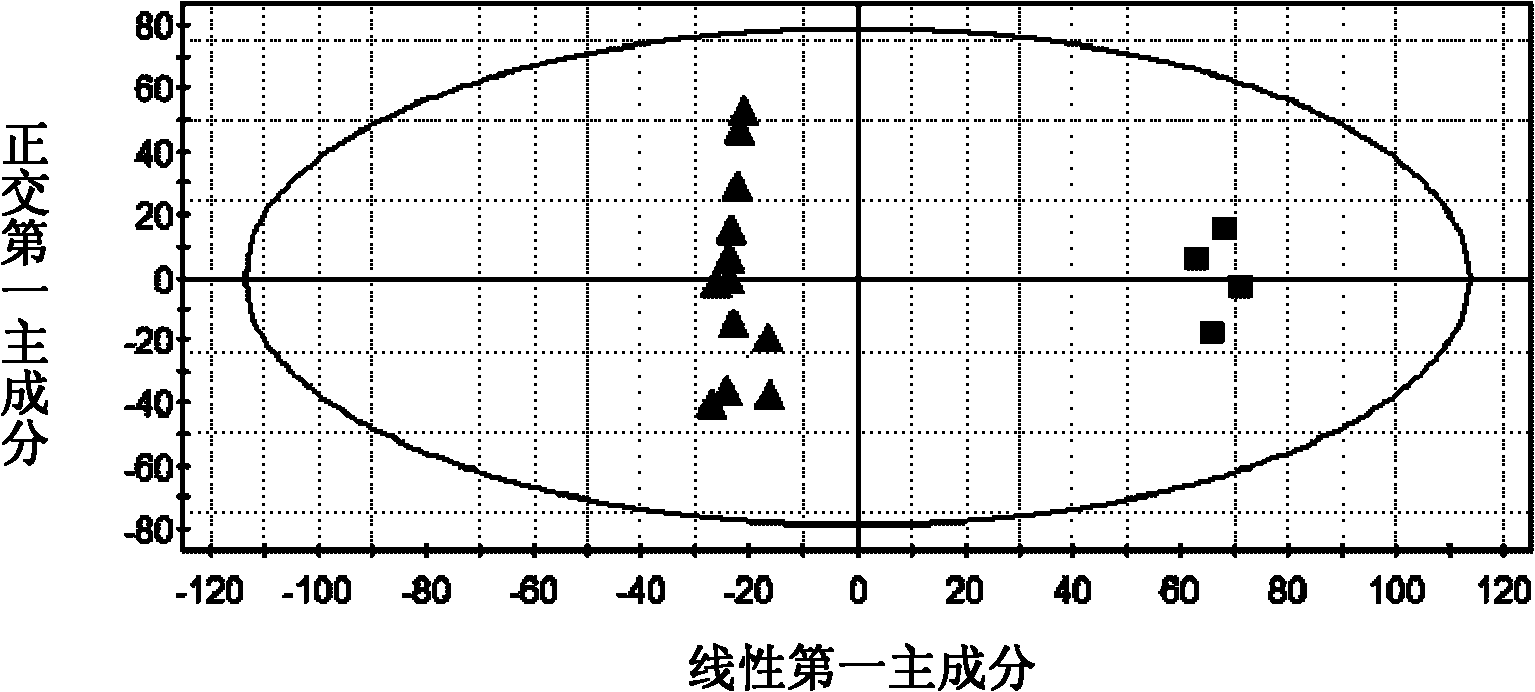
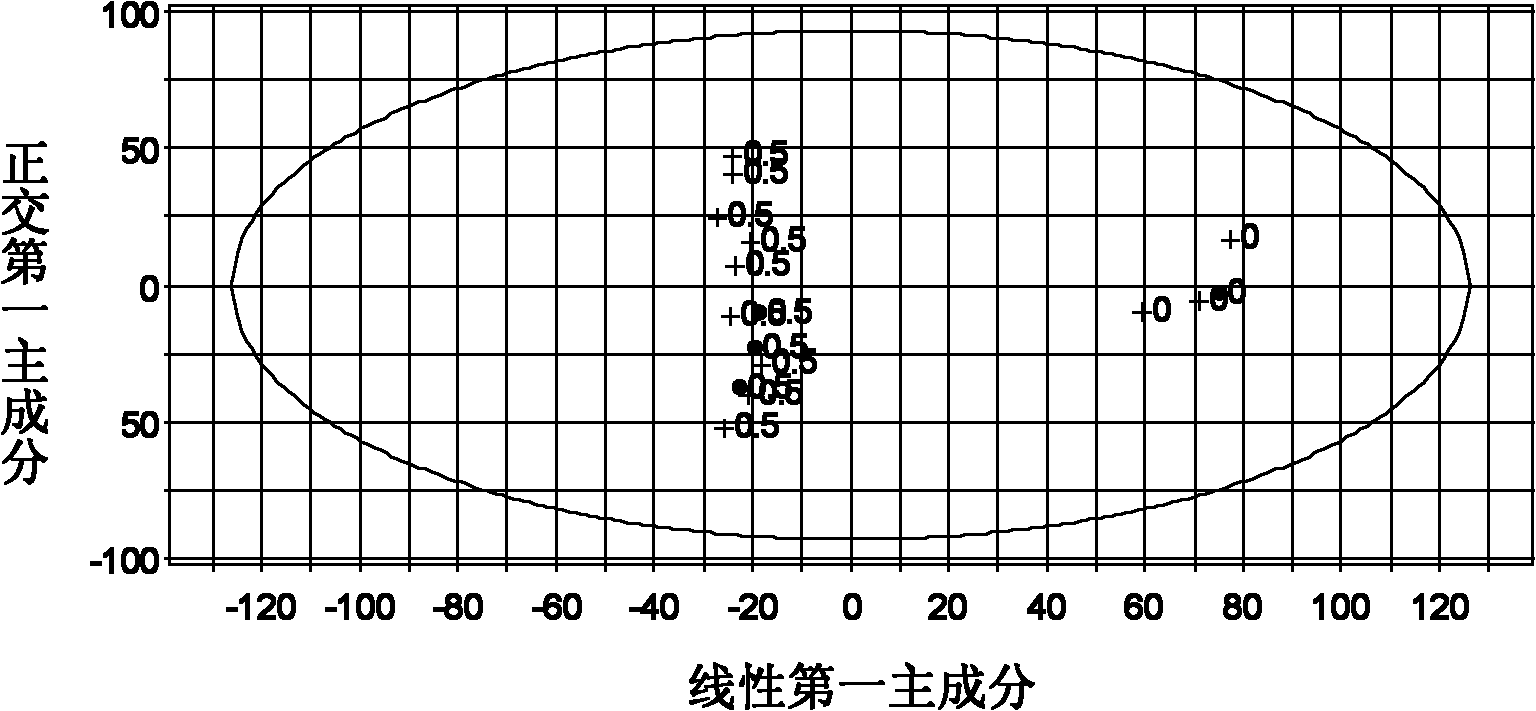
A method to discover and characterize lipid biomarkers in unicellular algae
A biomarker, unicellular algae technology, applied in the field of discovery and identification of unicellular algae lipid biomarkers, can solve the problems of inability to analyze thermally unstable metabolites, limited research scope, etc., to reduce the effect of ion inhibition , The effect of improving analytical throughput and high resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Discovery and Characterization of Lipid Biomarkers of Chlamydomonas nivalis Adaptation to NaCl Stress Environment 1, Chlamydomonas nivalis Cultivation and Harvesting
[0027]After Chlamydomonas nivalis was cultured for 6 days, a sterilized NaCl solution was added to make the final concentration 0.5% (w / v). Continue to cultivate for 0h, 1h, 7h, and 15h to take samples, and set four parallel samples at each time. The algae liquid was centrifuged (5000r / min, 5min), the supernatant was removed, the algae mud was washed three times with pure water, placed in an ultra-low temperature refrigerator for pre-freezing, and freeze-dried to obtain algae powder.
[0028] 2. Extraction of cell lipid compounds
[0029] The algae powder was extracted with methanol-chloroform-water extract. First add 0.025% (w / v, g / ml) antioxidant BHT to 2.5ml of methanol-chloroform-water (1 / 1 / 0.5, v / v / v) extraction solution, and then accurately weigh 10.0mg Algae powder, add the extract, shake with a...
Embodiment 2
[0057] Discovery and Characterization of Lipid Biomarkers of Chlamydomonas nivalis Adaptation to NaCl Stress Environment 1, Chlamydomonas nivalis Cultivation and Harvesting
[0058] After Chlamydomonas nivalis was cultured for 6 days, a sterilized NaCl solution was added to make the final concentration 0.5% (w / v). Continue to cultivate for 0h, 1h, 7h, and 15h to take samples, and set four parallel samples at each time. The algae liquid was centrifuged (5000r / min, 5min), the supernatant was removed, the algae mud was washed three times with pure water, placed in an ultra-low temperature refrigerator for pre-freezing, and freeze-dried to obtain algae powder.
[0059] 2. Extraction of cell lipid compounds
[0060] The algae powder was extracted with methanol-chloroform-water extract. First add 0.05% (w / v, g / ml) antioxidant BHT to 2.5ml of methanol-chloroform-water (1 / 1 / 0.5, v / v / v) extraction solution, and then accurately weigh 10.0mg Algae powder, add the extract, shake with a...
Embodiment 3
[0073] Discovery and Characterization of Lipid Biomarkers of Chlamydomonas nivalis Adaptation to NaCl Stress Environment 1, Chlamydomonas nivalis Cultivation and Harvesting
[0074] After Chlamydomonas nivalis was cultured for 6 days, a sterilized NaCl solution was added to make the final concentration 0.5% (w / v). Continue to cultivate for 0h, 1h, 7h, and 15h to take samples, and set four parallel samples at each time. The algae liquid was centrifuged (5000r / min, 5min), the supernatant was removed, the algae mud was washed three times with pure water, placed in an ultra-low temperature refrigerator for pre-freezing, and freeze-dried to obtain algae powder.
[0075] 2. Extraction of cell lipid compounds
[0076] The algae powder was extracted with methanol-chloroform-water extract. First add 0.05% (w / v, g / ml) antioxidant BHT to 2.5ml of methanol-chloroform-water (1 / 1 / 0.5, v / v / v) extraction solution, and then accurately weigh 10.0mg Algae powder, add the extract, shake with a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com