Method for detecting DNA single base mutant color by using nuclease reaction
A single-base mutation and color detection technology, applied in the field of DNA single-base mutation color detection, can solve the problems of reduced stability, difficult detection, cumbersome experimental steps, etc., and achieve the effect of improving selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Preparation of unmodified gold nanoparticles.
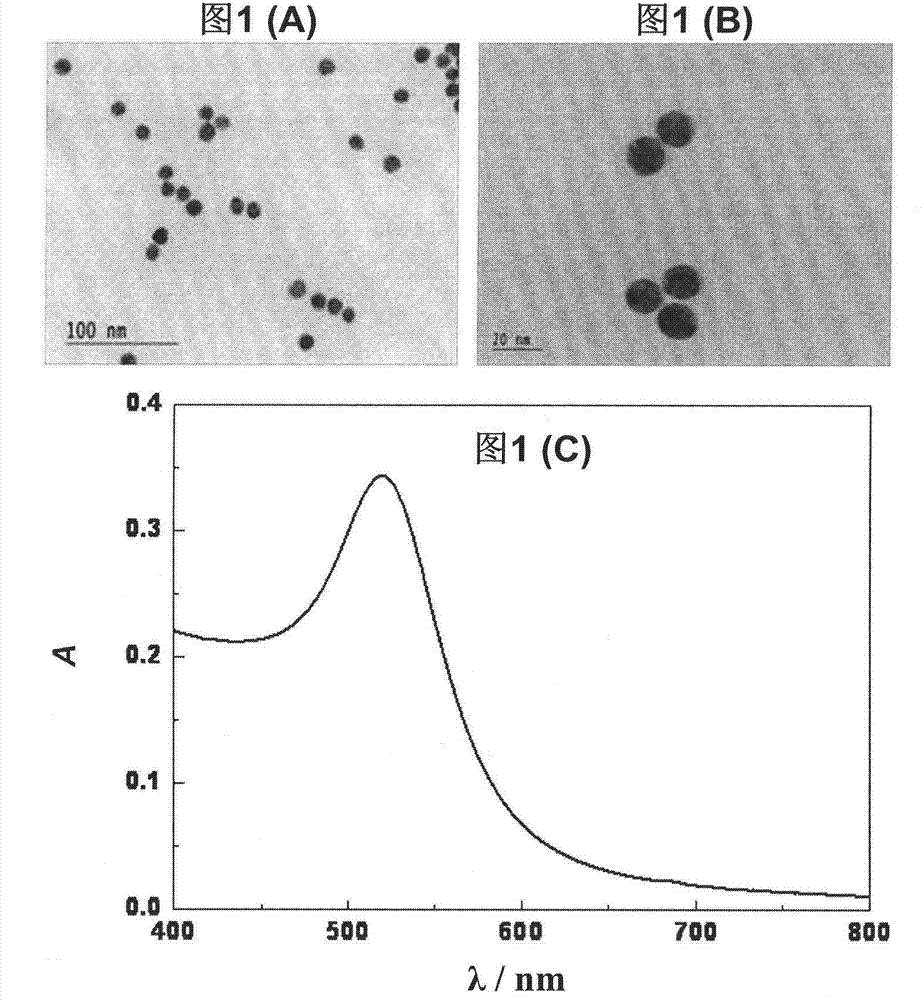
[0056] An aqueous solution of trisodium citrate (25ml, 38.8mM) was quickly added to a boiling auric acid HAuCl4 solution (250ml, 1mM). After a few minutes, the color of the solution changed from light yellow to dark red. The solution continued to reflux and stir for 15 min to complete the reaction. Then cool slowly to room temperature. Store at 4°C. According to the UV absorption intensity of gold nanoparticles at 520nm ( figure 1 (C)), the concentration of prepared nano-gold is 12 ± 1nM, and the size is 13nm (see figure 1 (A) and (B)).
Embodiment 2
[0057] Example 2: Differences in the stabilizing effects of dNMP, single-stranded DNA and double-stranded DNA on unmodified gold nanoparticles
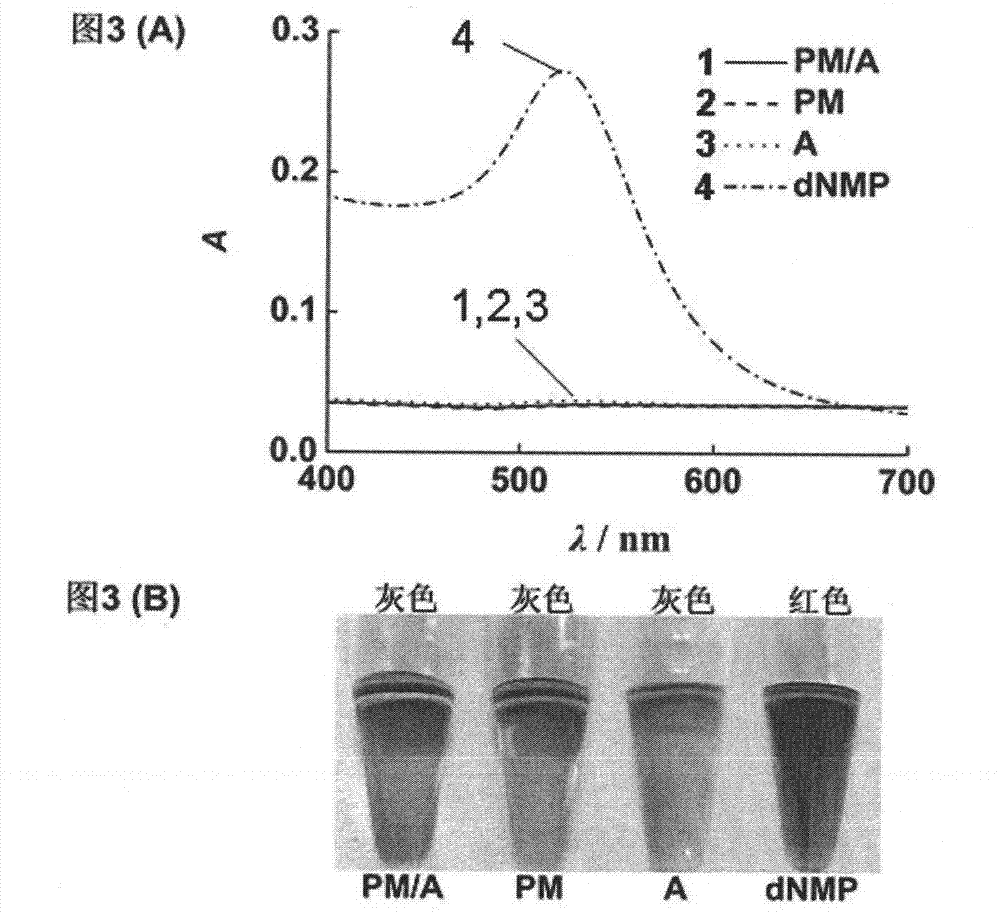
[0058] The purpose of this example is to confirm that dNMP, the nuclease degradation product of nucleic acid, can stabilize gold nanoparticles better than single-stranded and double-stranded DNA, which is the basis of the present invention.
[0059] Double-stranded DNA sample PM / A: 0.25 microliters of 100 μM single-stranded DNA probe A (see Table 1), 0.25 microliters of 100 μM target DNA PM that is completely complementary to single-stranded DNA probe A (see Table 1), 1 µl of buffer (0.6M NaCl, 10 mM phosphate, pH 7.4), and 0.5 µl of water, and kept at room temperature for 10 minutes.
[0060] Single-stranded DNA sample PM: composed of 0.25 μl of 100 μM single-stranded DNA probe PM, 1 μl of buffer (0.6M NaCl, 10 mM phosphate, pH 7.4), and 0.75 μl of water, and stored at room temperature 10 minutes.
[0061] ssDNA sample A: Consists ...
Embodiment 3
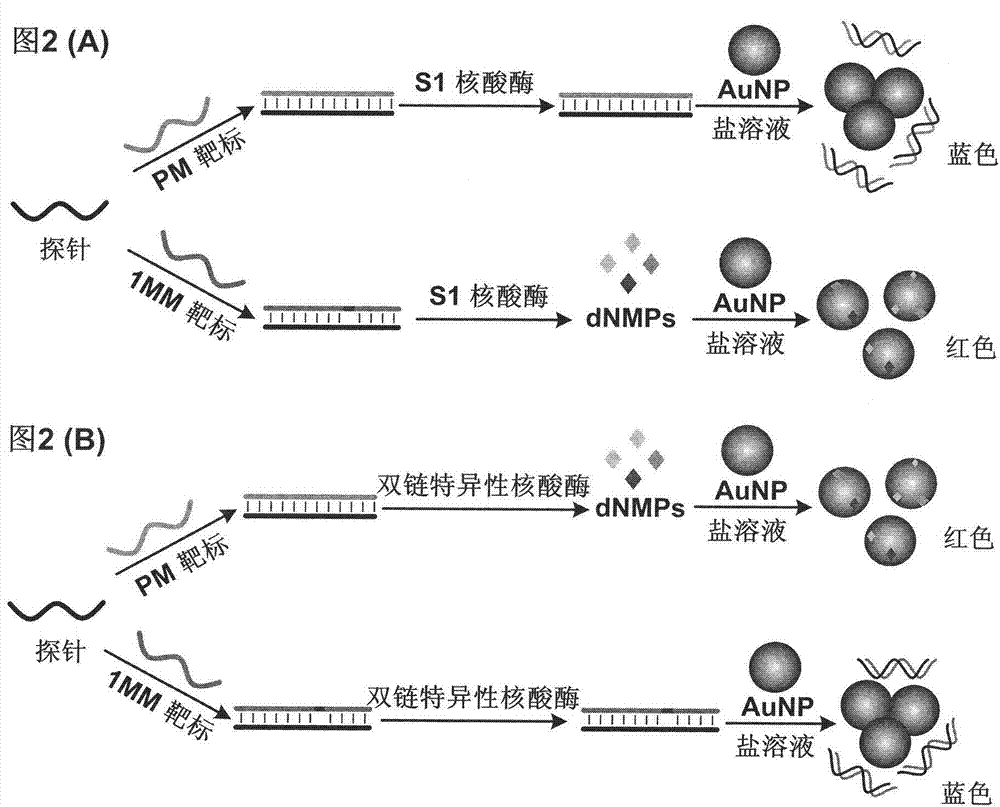
[0064] Example 3: The use of S1 nuclease for color detection of gold nanoparticles for single base mutations.
[0065] Reagent preparation: Prepare the solutions of probe A (Table 1), target PM (Table 1), target AC (Table 2), and target N (Table 1) with ultrapure water, and calibrate with a UV-visible spectrophotometer The concentration of the probe and each target solution was 100 μM. Prepare 5U μL with ultrapure water -1 S1 nuclease solution.
[0066] Take 1 μL each of probe A, target PM, AC or N, and mix with 1 μL 10× enzyme reaction buffer (containing 300 mM CH 3 COONa, 2800mM NaCl, and 10mM ZnSO 4 , pH 4.6) and 6 μL of ultrapure water were mixed and reacted at room temperature for 5 min. Take 1μL 5U μL -1 The S1 nuclease solution was added to the above mixture, mixed evenly, and reacted at 30°C for 30min. Take 2.5 μL of the above enzyme reaction mixture, add 50 μL of nano-gold solution with a concentration of 12±1 nM, and immediately add 40 μL of 0.5M NaCl phosphate...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com