Primer, kit and method for differentiating fins of different spieces of sharks by polymerase chain reaction and restriction fragment length polymorphism analysis (PCR-RFLP)
A technology of PCR-RFLP and kits, applied in biochemical equipment and methods, microbe determination/testing, DNA/RNA fragments, etc., to achieve high-sensitivity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] In this example, universal primers for different species of sharks were designed based on the mitochondrial 16S rDNA sequences of Sharkia and teleosts.
[0033] The inventors of the present invention have conducted a comprehensive analysis of the mitochondrial 16S rDNA gene sequences of different species of sharks and bony fishes in the NCBI database Shark class, based on GeneBank numbers AB015962.1, AJ310141.1, FJ853422.1, Y18134.1 Conserved sequences were compared by ClustalW software to find out the same sequence of their conserved sequences, so as to design corresponding primers.
[0034] After multiple comparisons, tests and verifications, a large number of primer pairs were screened to obtain the following two pairs of primers: The first round of primers:
[0035] SK-F11: CTCAGCCATCTTACCTGTGGCAAT (SEQ ID No. 1)
[0036] SK-R11 CYCCTCCTGCTGGGTCAAAG (SEQ ID No. 2)
[0037] Second round of primers:
[0038] Sharkfin-F2: CTACAAACCACAAAGATATCGGCACC (SEQ ID No. 3)
...
Embodiment 2
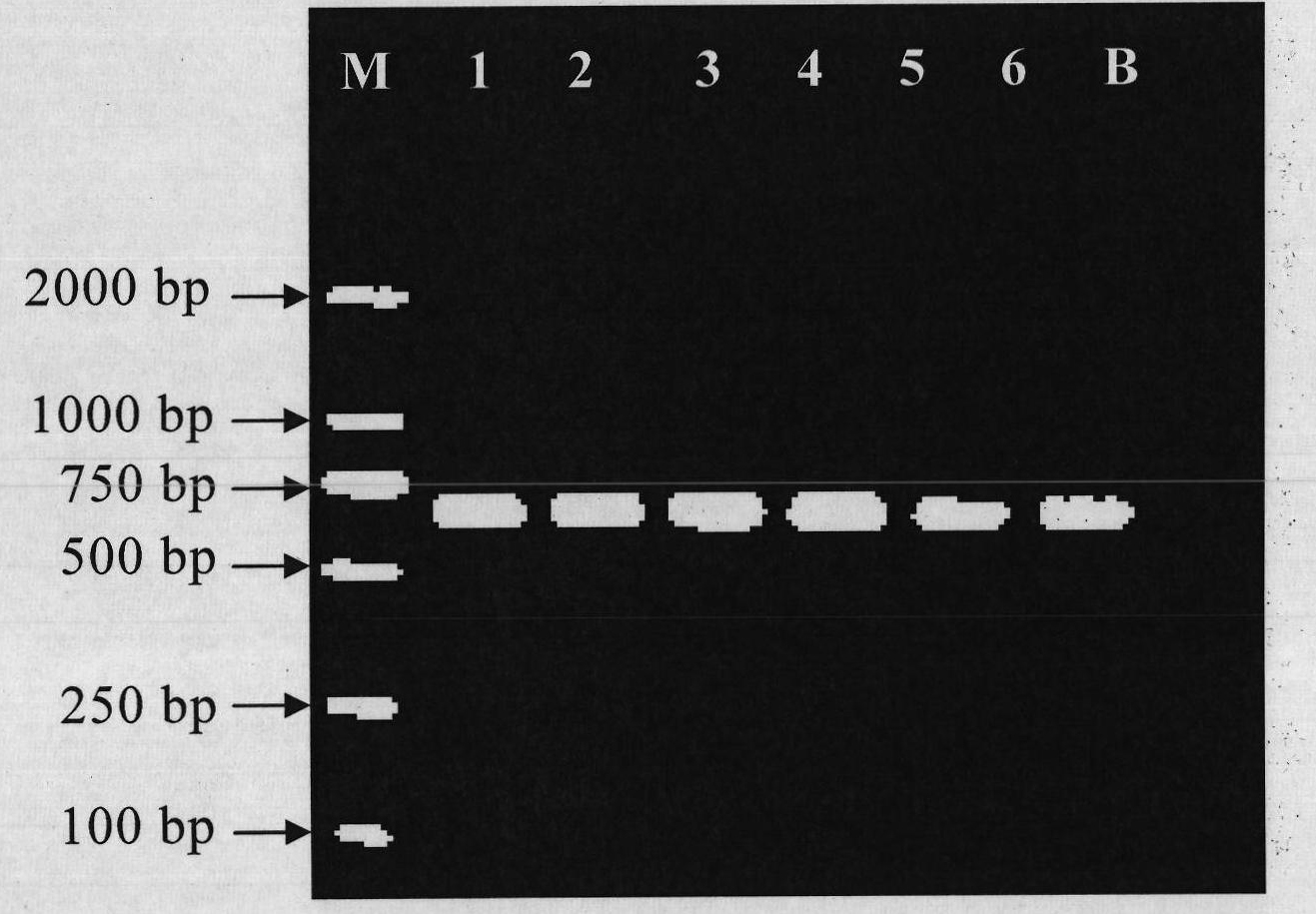
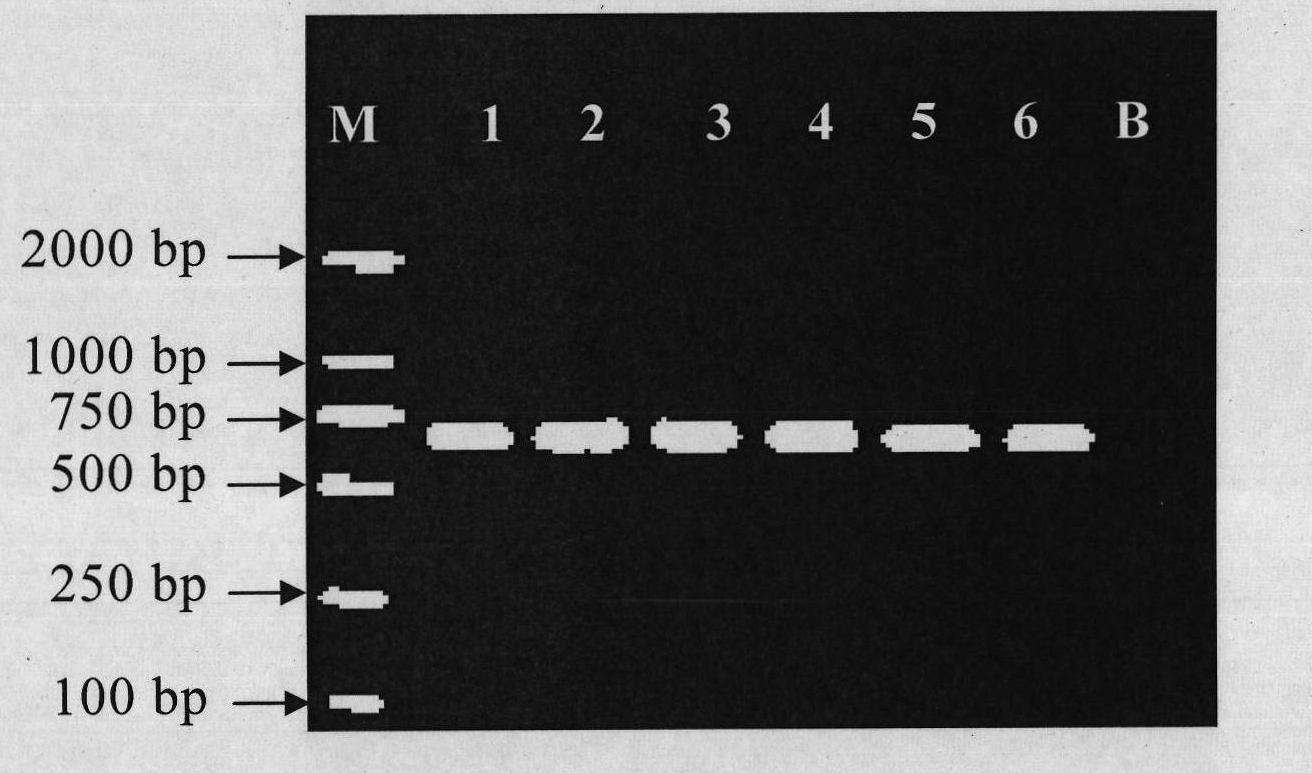
[0043] Nested PCR reactions were performed using universal primer pairs for sharks and teleosts. After each round of PCR, the quality of the total DNA extracted from the sample was tested to provide a suitable template for subsequent RFLP analysis.
[0044] The nest PCR reaction primer pair sequence that is used to detect Sharkidae and bony fish used in the present embodiment is:
[0045] The first round of primers consisted of SEQ ID No.1 and SEQ ID No.2, and the second round of primers consisted of SEQ ID No.3 and SEQ ID No.4.
[0046] By detecting the mitochondrial 16S rDNA sequence, test the extraction quality of the total DNA of the sample and provide a suitable template for the enzyme digestion reaction. The reaction system of Nest PCR in this example is: the first round of PCR reaction conditions is 94°C pre-denaturation for 8 minutes; 94°C for 30s, 50°C for 30s, 72°C for 60s, 35 cycles, and 2% agarose electrophoresis after the reaction Analysis; the second round of PC...
Embodiment 3
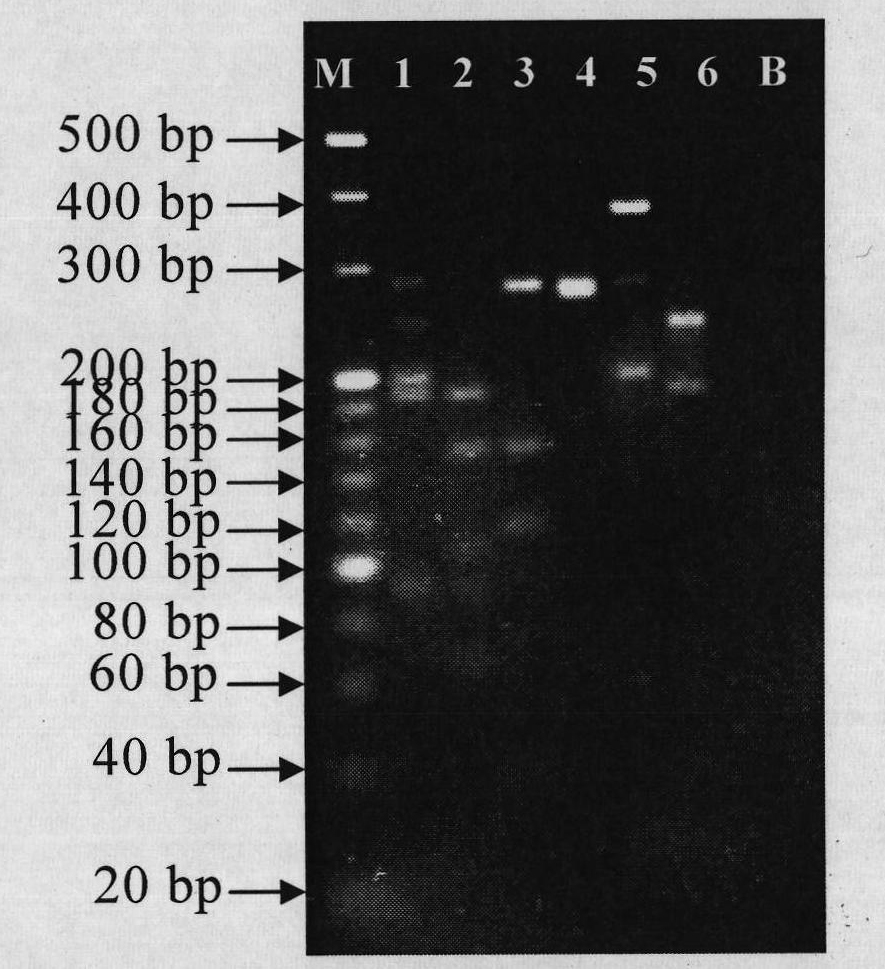
[0066] In this embodiment, PCR-RFLP analysis is performed on shark samples through the following experiments.
[0067] The digested product used in this example is the end product of the PCR reaction obtained by the nested PCR method described in Example 2.
[0068] The specificity of the shark primers can be determined by digesting the mitochondrial 16S rDNA sequence. A total of 20 μL of the entire reaction system: ddH 2 O 7.5 μL, 10×NEB Buffer 2 μL, AluI 0.5 μL, PCR reaction product 10 μL. Water bath at 37°C for 2h; after enzyme digestion, inactivate the enzyme activity in a 65°C oven for 10min. After the reaction, 4% agarose electrophoresis analysis was performed.
[0069] The main detection instruments used:
[0070] Micropipette (10 μL, 100 μL, 1000 μL Eppendorf), high-speed desktop centrifuge (Pico17 Thermo), high-speed pulverizer (IKA-WEARKE GERMANY), nucleic acid and protein analyzer (DYY-6C Beijing Liuyi Instrument Factory), water bath, Ordinary PCR instrument, e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com