Kit and method for quickly detecting enterotoxin generating escherichia coli
A technology of Escherichia coli and detection method, which is applied in the field of kits for rapid detection of enterotoxin-producing Escherichia coli, and can solve the problems of complicated operation and long detection time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
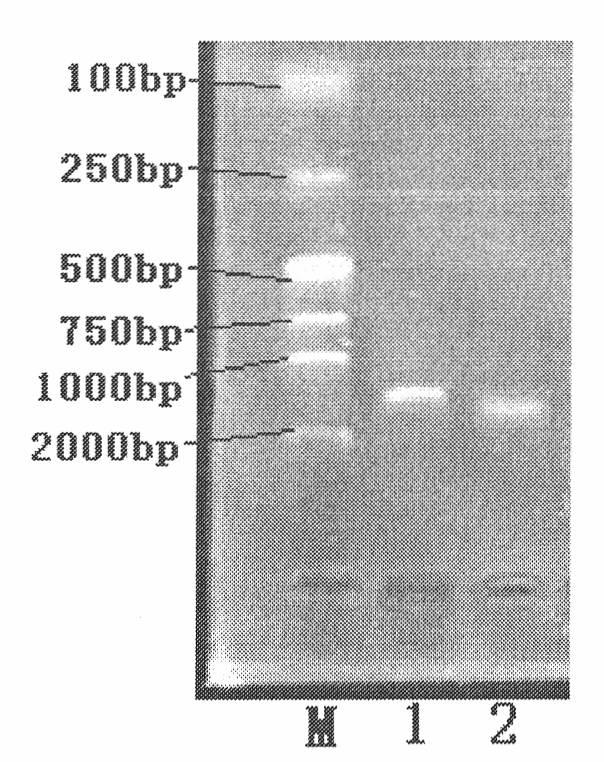
[0090] Embodiment 1 The primer specificity experiment of the present invention
[0091] Test materials and methods
[0092] 1 Culture of strains
[0093] (1) Take the preserved heat-labile enterotoxigenic Escherichia coli (C83902) and streak on the nutrient broth agar plate medium to isolate a single colony, and place it in an incubator at 37°C for upside-down culture for 24 hours.
[0094] (2) Pick a single colony and inoculate it into a 3ml liquid culture medium (test tube), put it into a shaker at 180rpm at 37°C for 12h.
[0095] (3) Transfer the bacteria cultured in the test tube into 120 ml of liquid culture medium and culture them under the same conditions for 12 hours, and store them at 4°C for later use.
[0096] 2 Genome Extraction
[0097] (1) Take 100 μl of bacterial culture and centrifuge at 12000 rpm for 5 minutes;
[0098] (2) Add 100 μl of sterile water, mix well, and place in a water bath at 100°C for 10 minutes, then in an ice bath for 2 minutes;
[0099]...
Embodiment 2
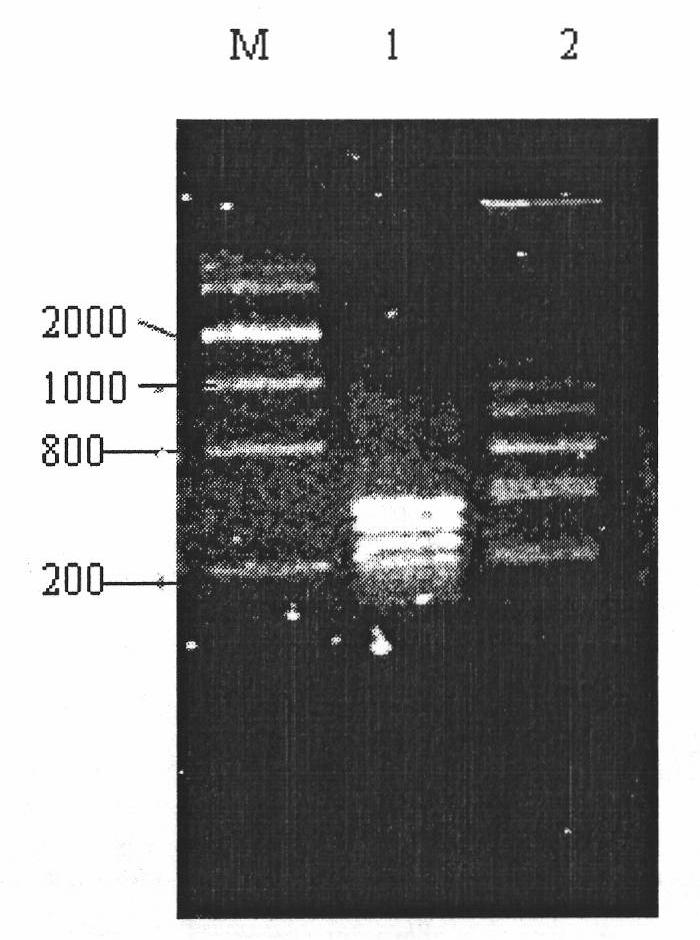
[0117] The sensitivity experiment of embodiment 2 primers of the present invention
[0118] According to the method of Example 1, the bacterial solution of different dilution concentrations is amplified with the primers of the present invention, and as the concentration of the bacterial solution decreases, the brightness of the specific band gradually becomes weaker until the bacterial solution is diluted to 10 -7 (the concentration dilution factor of the original bacterial solution) there are still bands, but the bacterial solution is diluted to 10 -8 When no amplification product appears (such as Figure 4 shown).
[0119] The detection sensitivity of the primer and method of the present invention is relatively high, and can be carried out in a relatively simple environment, and the requirements for samples are not high. The heat-labile enterotoxigenic Escherichia coli was directly lysed, and its suspension was collected for LAMP reaction. 3 μL of each reaction system solu...
Embodiment 3
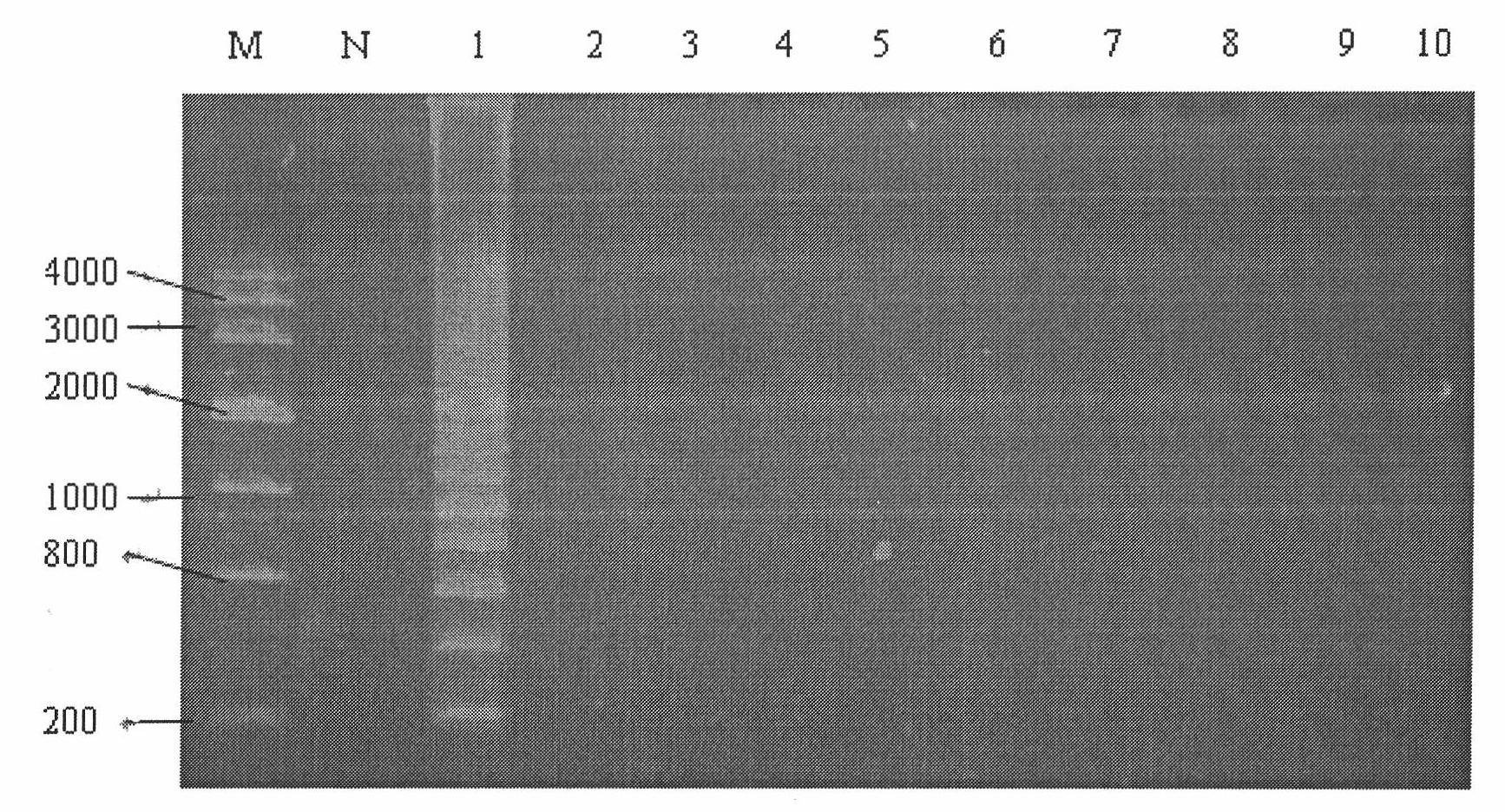
[0120] Embodiment 3 detects the preparation of enterotoxigenic Escherichia coli kit
[0121] Components: Specific Primers:
[0122] SEQ ID NO: 1 attacatttaagagcggcgc;
[0123] SEQ ID NO: 2 ggttcctagcattagacatgcttt;
[0124] SEQ ID NO: 3 gtgtatggaataataaaaccccctaaagcaaactagttttcca;
[0125] SEQ ID NO: 4 gtgtccttcatcctttcaatggcaggtcgaagtcccgggcagtc
[0126] Reaction system: 2 μl of 0.2 μM primers 1 and 2 (SEQ ID NO: 1 and SEQ ID NO: 2), respectively;
[0127] 1.6 μM Primers 3 and 4 (SEQ ID NO: 3 and SEQ ID NO: 4) are 2 μl respectively;
[0128] 2.5mM dNTPs 4μl;
[0129] 0.8mM Betaine 5μl;
[0130] 1X Thermopol buffer 2.5μl;
[0131] Intercalator SYBR GreenI 5μl (chromogenic solution)
[0132] Kit specification: 50T / box.
[0133] kit combination two
[0134] 0.2 μM primers 1 and 2 (SEQ ID NO: 1 and SEQ ID NO: 2) are 1.5 μl respectively;
[0135] 1.6 μM Primers 3 and 4 (SEQ ID NO: 3 and SEQ ID NO: 4) are 1.5 μl respectively;
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com