Transgenic plants with increased stress tolerance and yield
A technology of transgenic plants and high yield, applied in plant peptides, plant products, genetic engineering, etc., can solve problems such as crop yield limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
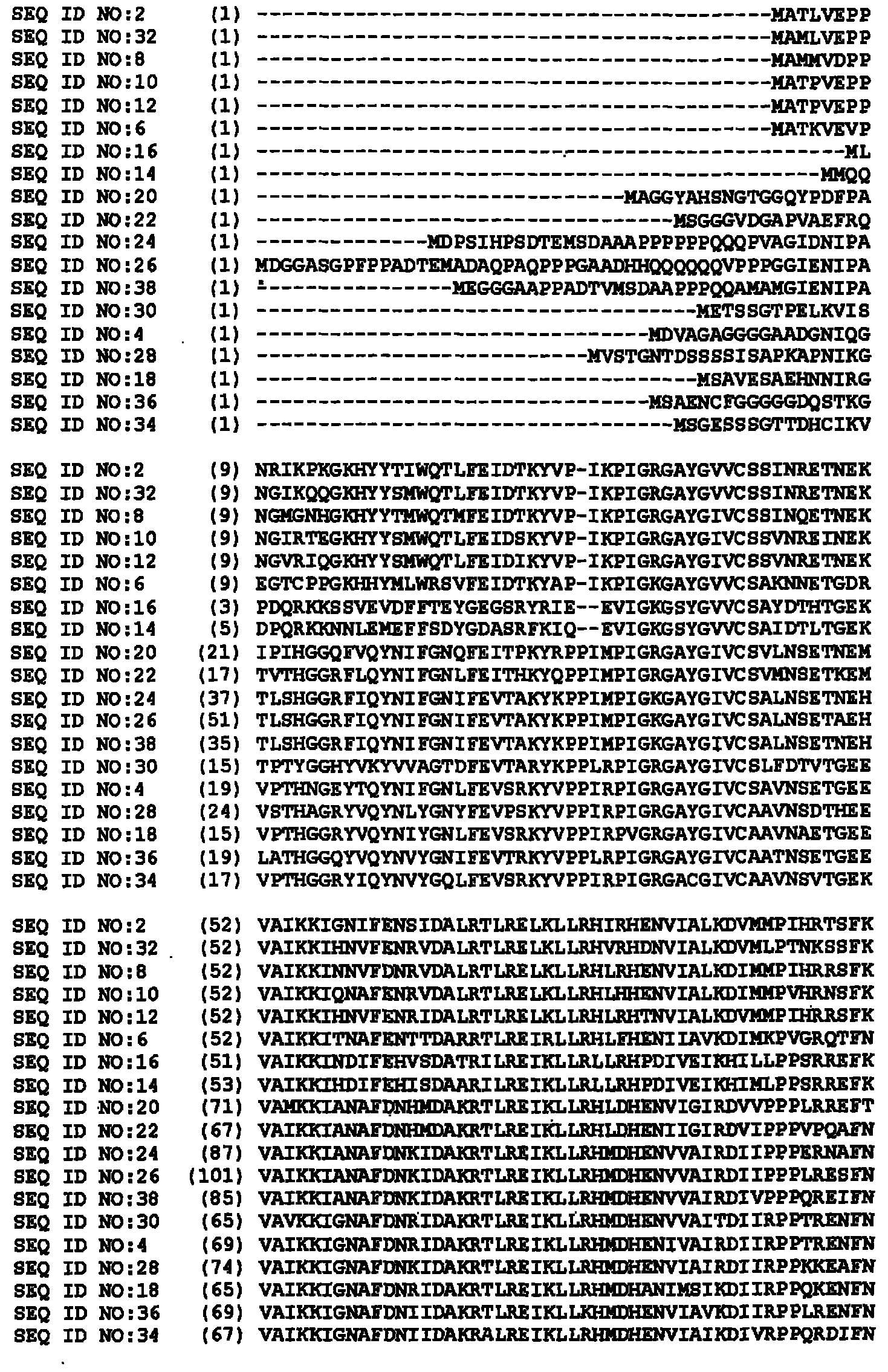
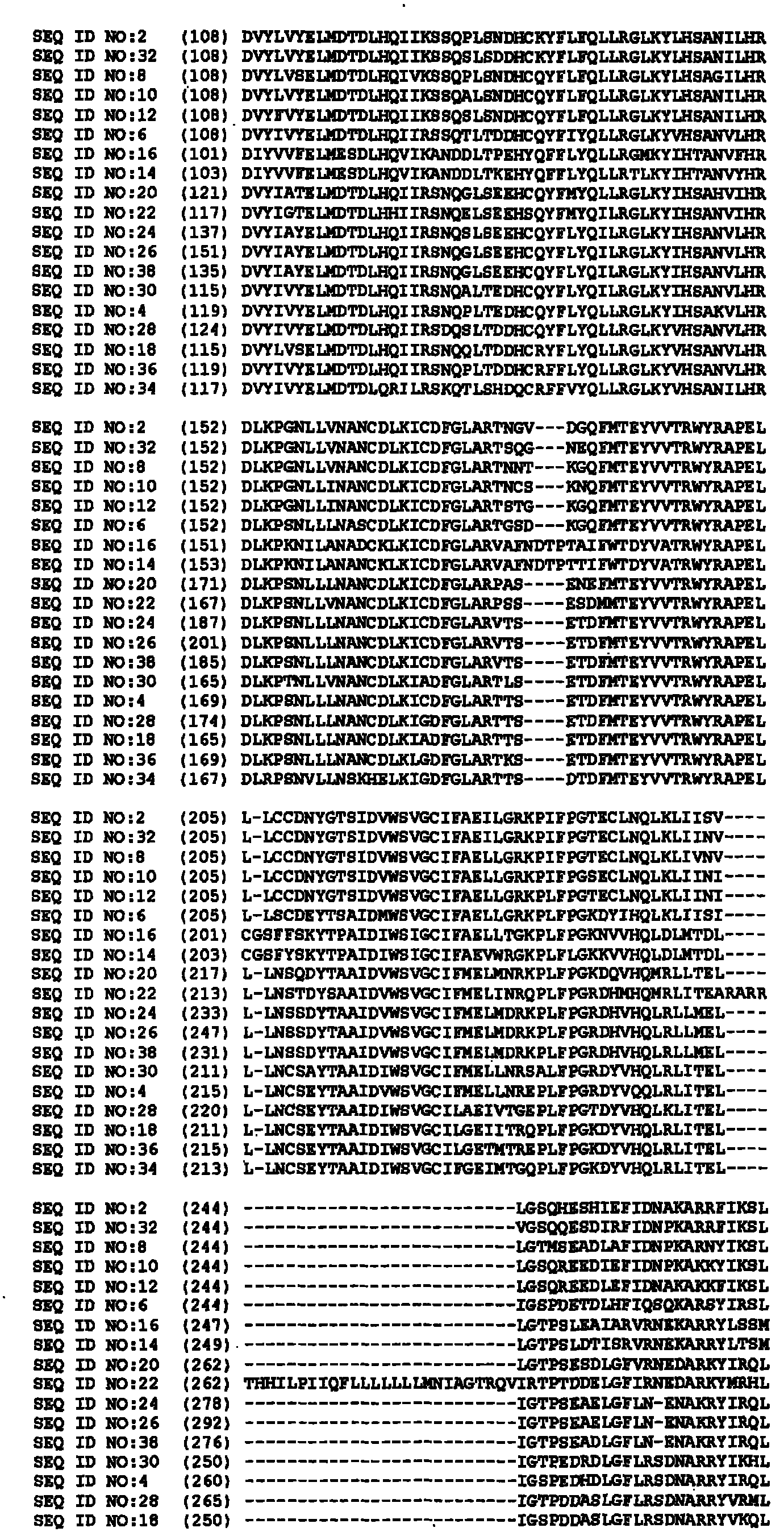
[0512] Characterization of cDNA
[0513] cDNAs are isolated from proprietary libraries of the corresponding plant species using known methods. Sequences were processed and annotated using bioinformatics analysis. The degrees of amino acid identity and similarity of the isolated sequences to their respective closest known public sequences are shown in Tables 2A to 11A, Tables 2B to 19B, Tables 2C to 16C, Tables 2D to 24D and Tables 2E to 4E Medium (using pairwise comparisons: gap penalty: 10; gap extension penalty: 0.1; scoring matrix: blosum62).
[0514] Table 2A
[0515] GM47143343 (SEQ ID NO: 2) compared to known mitogen-activated protein kinases
[0516] public database accession number
species
AAD32204
Apricot (Prunus armeniaca)
88.60%
NP_179409
85.90%
BAA04870
85.60%
CAN70944
Grapes (Vitis vinifera)
82.90%
ABO84371
Medicago truncatul...
Embodiment 2
[1009] gene characterization
[1010] Using standard recombination techniques, the lead genes (Lead gene) b1805 (SEQ ID NO: 287), YER015W (SEQ ID NO: 289), b1091 (SEQ ID NO: 317), b0185 (SEQ ID NO: 319), b3256 ( SEQ ID NO:321), b3255 (SEQ ID NO:329), b1095 (SEQ ID NO:335), b1093 (SEQ ID NO:343), slr0886 (SEQ ID NO:345) and slr1364 (SEQ ID NO:397 ). The functionality of each lead gene was predicted by comparing the amino acid sequence of each lead gene to other genes of known function. Homologue cDNAs were isolated from proprietary libraries of the corresponding species using known methods. Analyze, process and annotate sequences using bioinformatics. The degree of amino acid identity of the isolated sequences to their respective closest known public sequences (using pairwise comparisons: gap penalty: 10; gap extension penalty: 0.1; scoring matrix: blosum62) was used for the selection of homologous sequences, as shown in Tables 2F to 11F.
[1011] Form 2F
[1012] b1805 (...
Embodiment 3
[1054] gene characterization
[1055] The sterol pathway genes B0421 (SEQ ID NO:413), YJL167W (SEQ ID NO:415), SQS1 (SEQ ID NO:435) and YGR175C (SEQ ID NO:443) were cloned using standard recombinant techniques. The functionality of each sterol pathway gene was predicted by comparing the amino acid sequence of the gene with other genes of known function. Homologue cDNAs are isolated from proprietary libraries of the corresponding species using known methods. Analyze, process and annotate sequences using bioinformatics. The degree of amino acid identity of the isolated sequences to their respective closest known public sequences is shown in Tables 2G to 5G (using pairwise comparisons: gap penalty: 11; gap extension penalty: 1; scoring matrix: blosum62). The amino acid identity and degree of similarity of the isolated sequences to the respective closest known public sequences was used for the selection of homologous sequences as described below.
[1056] Form 2G
[1057] Comp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com