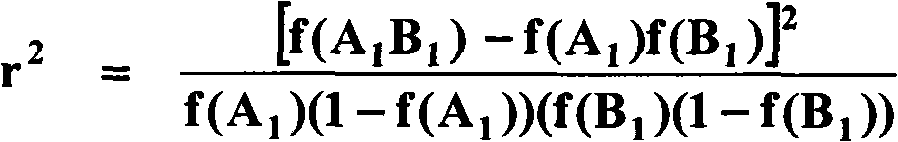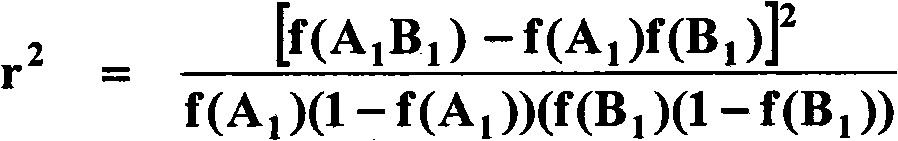Genetic markers for horned and polled cattle and related methods
A marker, associated technology, applied to genetic markers of horned and hornless cattle and related fields
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0109] Example 1 : Identification of genetic markers associated with horned / hornless phenotypes in dairy cows
[0110] Georges et al. used microsatellite markers in 1993 to locate the hornless mutation in Jersey cows at the proximal end of bovine chromosome 1 (BTA01). More recent work has refined the mapping of the hornless locus, including mapping of microsatellite markers (Schmutz et al. 1995; Brenneman et al. 1996; Harlizius et al. 1997; 2005) and the creation of a BAC-based physical map of the hornless regions of chromosomes (Wunderlich et al 2006). The chromosomal positions of the near-central gene ATP5O and the most distal gene KRTAP8 in the hornless region of the chromosome are equivalent to those of the published bovine genome compilation version 3.1 (www.hgsc.bcm.tmc.edu / projects / bovine / ) compared with the referenced resources 0.6Mb and 3.9Mb.
[0111] The aim of this work was to identify single nucleotide polymorphisms (SNPs) associated with the hornless trait by...
Embodiment 2
[0121] Example 2 : Using single nucleotide polymorphisms to improve offspring characteristics
[0122] To improve the average genetic merit of a defined trait in a population, one or more markers with a clear association with that trait can be used in the selection of breeding animals. In the case of each locus discovered, using animals with marker alleles (or haploids of multiple marker alleles) that are in population-wide linkage disequilibrium with the desired phenotype, over time Shifting increases the frequency of the allele (and phenotype) in the population, thereby increasing the average genetic merit of the trait in the population. This increased genetic advantage can be propagated to commercial populations to fully realize its value.
[0123] For example, a DNA testing program protocol or the production of semen through the use of DNA markers as described herein can greatly alter the frequency of the hornless allele in a given population. Testing bull semen in a p...
Embodiment 3
[0137] Example 3 : Identification of SNPs
[0138] A nucleic acid sequence contains a SNP of the present invention if the nucleic acid sequence contains at least 20 consecutive nucleotides comprising and / or adjacent to the polymorphisms described in Tables 1 and 2 and the Sequence Listing. Alternatively, in cases where the shorter contiguous nucleotide sequence is unique in the bovine genome, the polymorphism described in Tables 1 and 2 and the Sequence Listing or adjacent to the shorter contiguous Nucleotide fragments to identify the SNPs of the present invention. Typically, a SNP site is characterized by a consensus sequence comprising a polymorphic site therein, the position of the polymorphic site, and the various alleles at the polymorphic site. "Consensus sequence" refers to a DNA sequence constructed to be identical at each nucleotide position of an aligned cluster of sequences. These clusters are commonly used to identify SNPs and Indel (insertion / deletion) polymor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com