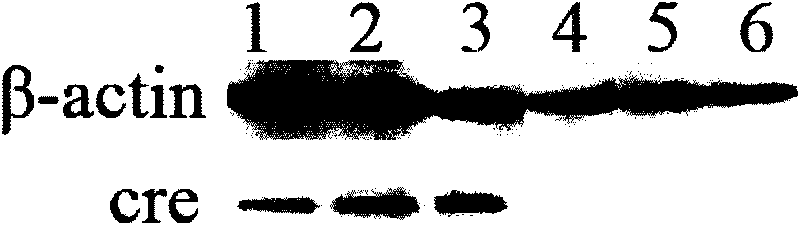Method for deleting exogenous gene in transgenic cell at fixed point
An exogenous gene and cell technology, applied in the field of genetic engineering, can solve the problems of increasing the difficulty of vector construction and cell screening steps, and achieve the effect of avoiding drug screening.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1c
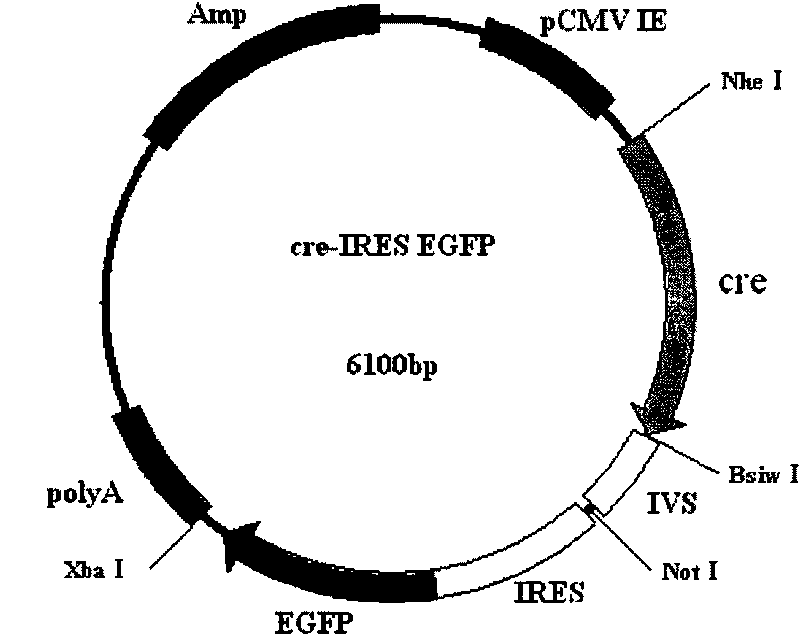
[0023] The construction of embodiment 1cre recombinase eukaryotic expression vector
[0024] Using the PBS 185 plasmid as a template, PCR primers were designed to amplify the CDS sequence of cre recombinase, and at the same time, NheI and BsiWI restriction sites were introduced at both ends of the sequence.
[0025] The designed upstream and downstream primers are:
[0026] Nhe I-Cre-F:GAA GCTAGC ATGTCCAATTTACTGACCGTAC
[0027] Bsi WI-Cre-R:TCT CGTACG CTAATCGCCATCTTTCCAGCAG
[0028] The reaction system for Cre CDS amplification is: 50 μl reaction system, including 1 μl of PBS185 plasmid (200 ng / μl), 1 μl of each primer (20 pmol / μl), 6 μl of dNTP (10 mM), 5 μl of 5×LA PCR Buffer, LA Taq enzyme (Takara ) 0.3 μl.
[0029] The reaction program was: pre-denaturation at 94°C for 5 min, denaturation at 98°C for 10 s, annealing and extension at 68°C for 1 min, a total of 35 cycles, and 10 min at 72°C.
[0030] The PCR product was recovered and connected to the pMD19-T vector. ...
Embodiment 2
[0032] Example 2 Transfection of 293T cells and bovine fibroblasts and expression detection of cre recombinase
[0033] 2.1 Cell culture and transient transfection
[0034] Take bovine ear tissue and cut into 1mm 3 For the left and right small pieces, wash them twice with DMEM / F12 and then plant them in batches in a 25cm medium containing 1mL DMEM / F12+10%FBS. 2 In the culture flask, add DMEM / F12+10% FBS to 6mL after the tissue block is firmly adhered to the wall, and store at 37°C, 5% CO 2 Cultivate in an incubator for 6-7 days, change the medium once every 2 days, after the cells grow confluent, digest and passage with 0.25% trypsin for 2-3 times, freeze in batches with DMEM / F12+20%FBS+10%DMSO, and culture Bovine fibroblasts.
[0035] 293T cells and bovine fibroblasts were respectively inoculated in 6-well cell culture plates, and when they were cultured in DMEM medium containing 10% fetal bovine serum to 70-80% confluence, according to the instructions, Lipofectamine 2000...
Embodiment 3
[0038] Example 3 Screening and identification of gene deletion cells
[0039] 1LoxP-N n -Construction of the LoxP structure
[0040] The two ends of the Neo expression frame of the two LoxP site primers pIRES-NEO vector were constructed by primer amplification or restriction enzyme connection to form pIRES-LoxP-NEO-LoxP, and the two ends of the LoxP-NEO-LoxP structure can be introduced into special Restriction sites for connection to other specific expression vectors. The NEO gene in the LoxP-NEO-LoxP structure can also use any other gene structure (N n , N represents the nucleotide sequence, and n represents the number of nucleotides) to replace, that is, to construct LoxP-N n -LoxP structure.
[0041] 2 Cell culture and transient transfection
[0042] The above LoxP-N n -LoxP structure (take the LoxP-neo-LoxP structure as an example) to directly transfect bovine fibroblasts to obtain transgenic bovine fibroblasts that stably integrate the LoxP-neo-LoxP structure, and s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com