Polygonatum filipes microsatellite markers
A technology of microsatellite markers and Polygonatum, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of poor repeatability, late start of molecular marker research, and inability to distinguish pure and genotypes from heterozygous genes Type and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] 1. Construction of Polygonatum long stem microsatellite DNA enrichment library
[0027] 1.1 Genome extraction and digestion:
[0028] Using the CTAB method introduced by Doyle J (Doyle J. DNA protocols for plants-CTAB totalDNA isolation [A]. In Hewitt GM, Johnston A (eds.), Molecular Techniques in Taxonomy [M]. Berlin: Springer, 1991, 283-293 ) to extract the genome, digest the genome with the restriction endonuclease Sau3AI (purchased from TaKaRa company), and electrophoresis the digested product on 1% agarose gel, cut out the DNA fragment of 300-900bp size under ultraviolet light and use the gel recovery kit (purchased from Shanghai Sangong) to purify and recover the digested product.
[0029] 1.2 Add connector:
[0030] Add the two ends of the purified and recovered enzyme-cut product to the adapter:
[0031] LA(5'-GGCCAGAGACCCCCAAGCTTCG-3')
[0032] LB(5'-PO 4 -GATCCGAAGCTTGGGGTCTCTGGCC-3'),
[0033] Under the action of T4 ligase (purchased from Promega), the ...
Embodiment 2
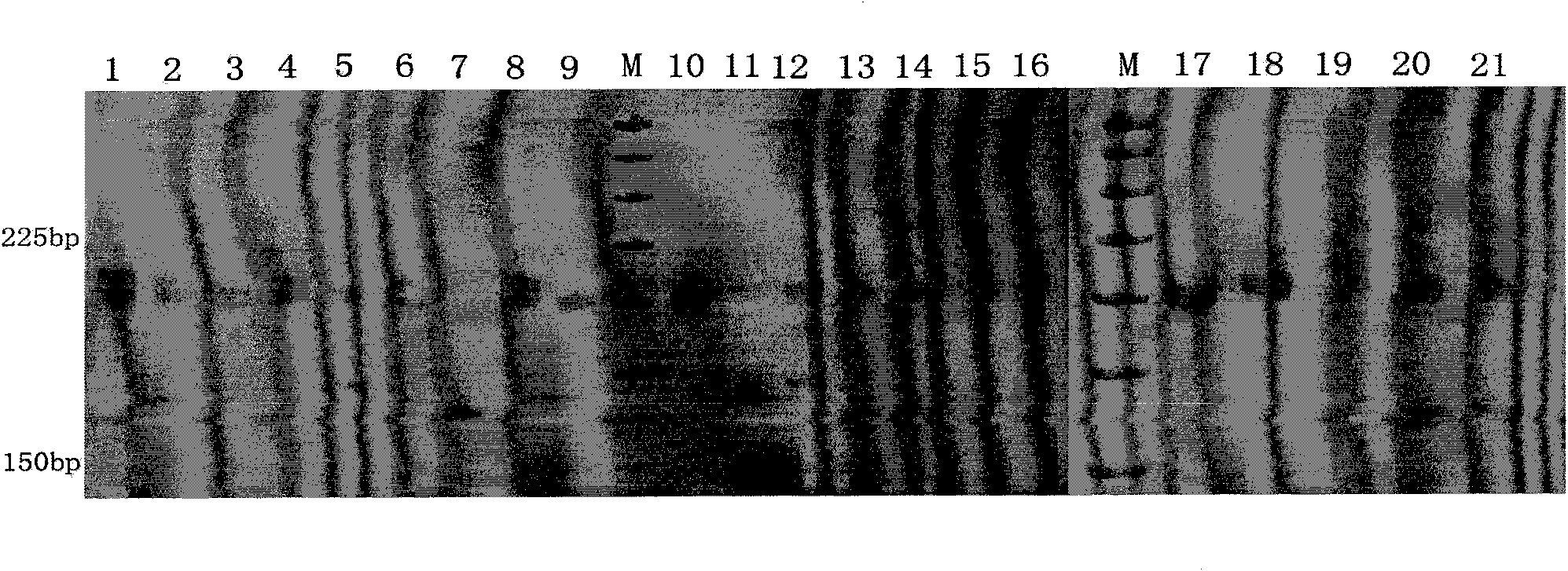
[0052] 2.1 Screen polymorphic primers:
[0053] Use the CTAB method of 1.1 to extract the Rhizoma Polygonatum genome. The genome was amplified with designed primers (Table 1). Reaction program: pre-denaturation at 94°C for 4 minutes, denaturation at 94°C for 45 seconds, annealing for 30 seconds (see Table 1 for annealing temperature), extension at 72°C for 35 seconds, and three steps from denaturation to annealing were repeated 35 times. A final full extension at 72°C for 8 minutes. 1.5% agarose gel electrophoresis detection. PCR products were detected by PAGE electrophoresis, and a total of 11 polymorphic microsatellite markers were obtained; Pt1 was 250 nucleotides, Pt2 was 370 nucleotides, Pt3 was 277 nucleotides, and Pt4 was 905 nuclei Pt5 is 729 nucleotides, Pt6 is 343 nucleotides, Pt7 is 247 nucleotides, Pt8 is 403 nucleotides, Pt9 is 297 nucleotides, Pt10 is 298 nucleotides , Pt11 is 319 nucleotides.
[0054] 2.2 Analysis of results:
[0055] The expected heterozy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com