Molecular tagged method of indicating and identifying size of qin-chuan cattle body by using GDF5 gene
A technology of molecular markers and genes, applied in the field of animal molecular genetics, can solve the problems of hindered bone growth in limbs, affecting the body size, etc., and achieves the effects of high accuracy, significant economic benefits, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] Implementation example 1: Correlation between bovine GDF5 gene polymorphism and body size traits of Qinchuan cattle population
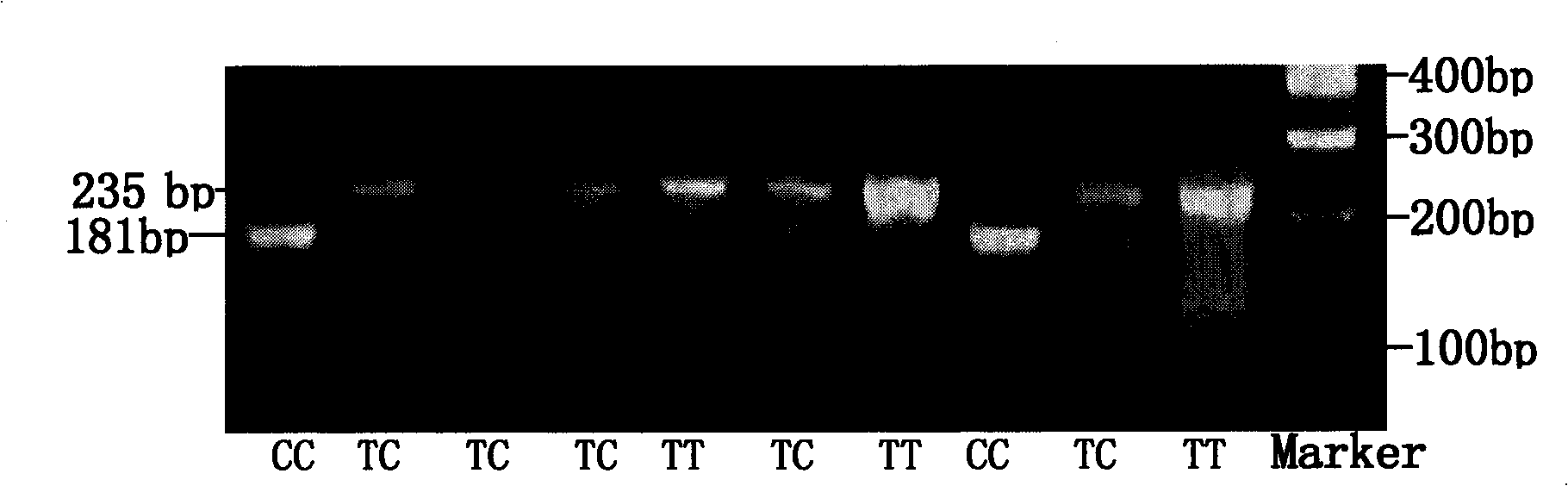
[0115] Utilize the primers (GDFF and GDFR) of the present invention to carry out PCR amplification to the genomic DNA of 393 individuals of Qinchuan cattle population, and utilize Mva I restriction endonuclease to digest the PCR product, then carry out 2.5% agarose gel electrophoresis analysis. Three genotypes were detected in the Qinchuan cattle population. When the mutation site in exon 1 of the bovine GDF5 gene is a C base, a restriction endonuclease Mva I restriction site (CC^TGG) will be generated. , Mva I will produce two fragments (181bp and 54bp) after digesting the PCR product, which will be named CC genotype; when this site is a T base (CTTGG), there is no Mva I restriction site, After digesting the PCR product, there is only 1 fragment (235bp), which is named as TT genotype (wild type); when this site is T / C heterozygous, Mva I will p...
Embodiment 2
[0127] Implementation example 2: Correlation between bovine GDF5 gene polymorphism and Qinchuan cattle improved population size traits
[0128] Utilize the primers (GDFF and GDFR) of the present invention to carry out PCR amplification to the genomic DNA of 240 individuals of Qinchuan cattle improved population, and utilize Mva I restriction endonuclease to digest the PCR product, then carry out 2.5% agarose gel electrophoresis analysis . Three genotypes were detected in the improved population of Qinchuan cattle. When the mutation site in exon 1 of the bovine GDF5 gene is C base, the restriction endonuclease Mva I restriction site (CC^TGG ), Mva I will produce two fragments (181bp and 54bp) after digesting the PCR product, which will be named CC genotype; when this site is a T base (CTTGG), there is no Mva I restriction site, After I digested the PCR product, there was only one fragment (235bp), which was named TT genotype (wild type); when this site was T / C heterozygous, Mv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com