Nucleic acid sensor based on quantum dots and preparation method and detection method thereof
A technology of nucleic acid sensors and quantum dots, which is applied in the field of biosensors, can solve the problems that have not yet been reported on nucleic acid sensors, and achieve the effect of repeated detection synchronization, simple preparation, and rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
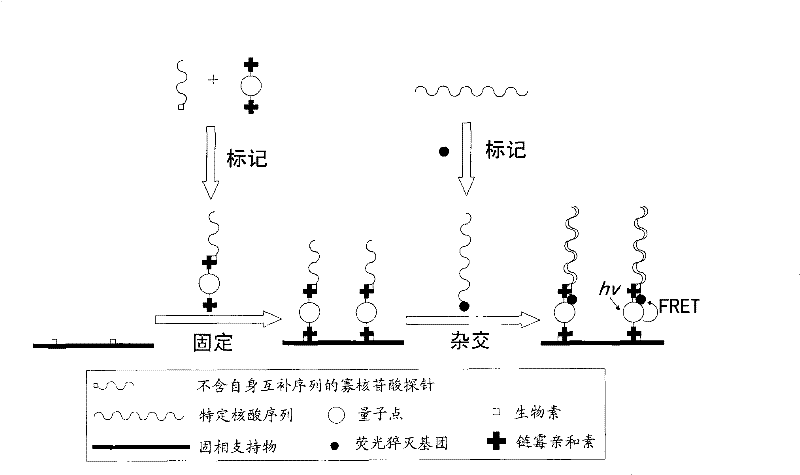
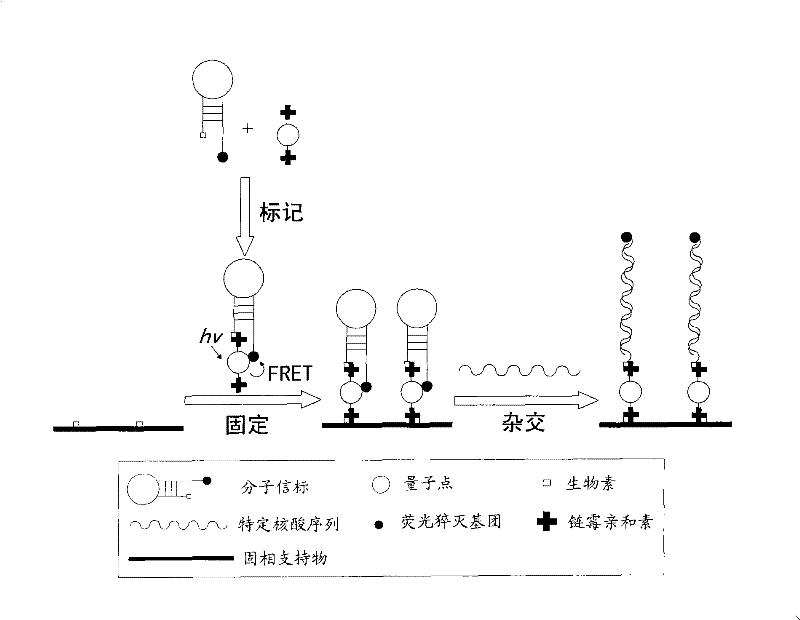
[0047] Schematic diagram of the preparation and detection of the first quantum dot-based nucleic acid sensor figure 1 As shown, the preparation of the nucleic acid sensor is to firstly couple the oligonucleotide without self-complementary sequence to the quantum dot through the biotin-avidin (avidin or streptavidin) system to prepare one end Quantum dot-labeled oligonucleotide probes; then quantum dots and solid phase supports (for nucleic acid sensors, optional solid phase supports include silicon wafers, glass slides, tiles, polypropylene membranes or nylon membranes, etc.) ) is coupled through a biotin-avidin (avidin or streptavidin) system, thereby immobilizing the oligonucleotide probes on the surface of the solid phase support through quantum dots; using the same method to immobilize multiple oligonucleotide probes to form an oligonucleotide probe array; when the nucleic acid sensor is used for detection, one end of the nucleic acid sample to be tested is labeled with a ...
Embodiment 1
[0054] 1. Preparation of nucleic acid sensor
[0055] 1. Preparation of streptavidin-labeled quantum dots
[0056] (1) Preparation of quantum dots
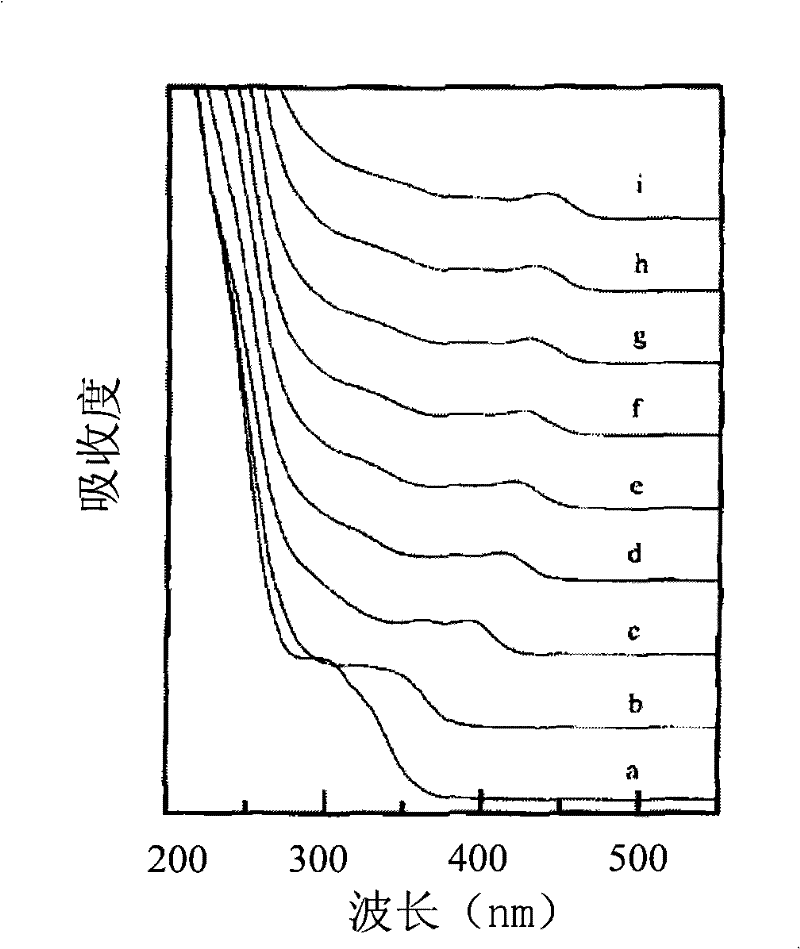
[0057] Mix sodium sulfate and selenium powder in a molar ratio of 3:1, add 150 mL of deionized water, and reflux reaction under nitrogen protection conditions until the selenium powder is completely dissolved to obtain a clear sodium selenosulfate solution; 0.46 g of cadmium chloride Dissolve in 240mL of deionized water, stir and react for 30 minutes under nitrogen protection, then add 0.36mL of thioglycolic acid dropwise, adjust the pH value of the solution to 11.2 with NaOH with a concentration of 1mol / L, continue to stir and react for 30 minutes, then drop Add 12 mL of the above-mentioned sodium selenosulfate solution, and by controlling the time of the reflux reaction, a CdSe quantum dot solution with an emission wavelength of 492 nm and a carboxyl group modified on the surface is prepared.
[0058] The UV-Vis absorption spe...
Embodiment 2
[0075] 1. Preparation of nucleic acid sensor
[0076] 1. Preparation of streptavidin-labeled quantum dots
[0077] (1) Preparation of quantum dots
[0078] Na 2 SeSO 3 Solution preparation: In a 250mL three-necked bottle, add Na 2 SO 3 6.302g (0.05mol) and Se powder 1.579g (0.02mol), add high-purity water 100mL to dissolve, stirring, N 2 Under protective conditions, heat up to 90°C for reflux reaction. As the reaction time prolongs, the solution containing gray-black precipitate gradually becomes clear and transparent. After 6 hours of reaction, stop heating and filter with suction to obtain a colorless and transparent filtrate, that is, the concentration is about 0.2mol / L. Na 2 SeSO 3 The solution is stored at a temperature of 70°C (to prevent oxidation) and is used for later use.
[0079] Preparation of NaHSe solution: In a 20mL small flask, add 0.1895g (0.0024mol) of Se powder, add 6mL of high-purity water to dissolve, and then add newly prepared NaBH 4 Solution [N...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com