Optimizing mass spectrogram model for detecting liver cancer characteristic protein and preparation method and application thereof
A technology of characteristic protein and mass spectrometry model, applied in the field of protein detection and mass spectrometry detection, can solve the problems of inability to detect low-abundance proteins, insufficient resolution, limited practical value, etc., to improve clinical cure, reduce fatality rate, and design accurate and reasonably practicable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Differentiation between normal and liver cancer patients caused by hepatitis B and preparation of mass spectrometry kits
[0052] (1) Experimental method
[0053] 66 patients with liver cancer caused by hepatitis B (including 15 cases of stage I, 18 cases of stage II, 16 cases of stage III, and 17 cases of stage IV, aged 43 to 70 years, with a median age of 50 years) and 24 cases of liver cancer caused by hepatitis B The preoperative serum of patients with liver cirrhosis (age 42-68 years, median age 45 years). The 90 control sera came from healthy volunteers (age 40-69 years old, median age 49 years old), and they came from the physical examination population with normal liver function and renal function tests. Collect 1mL of venous blood from the subject on an empty stomach, immediately after collection, let it stand in a refrigerator at 4°C for 2 hours, centrifuge at 4,000r / min at 4°C for 10 minutes to separate the serum, and centrifuge the serum again at ...
Embodiment 2
[0080] Embodiment 2 clinical trial and double-blind test
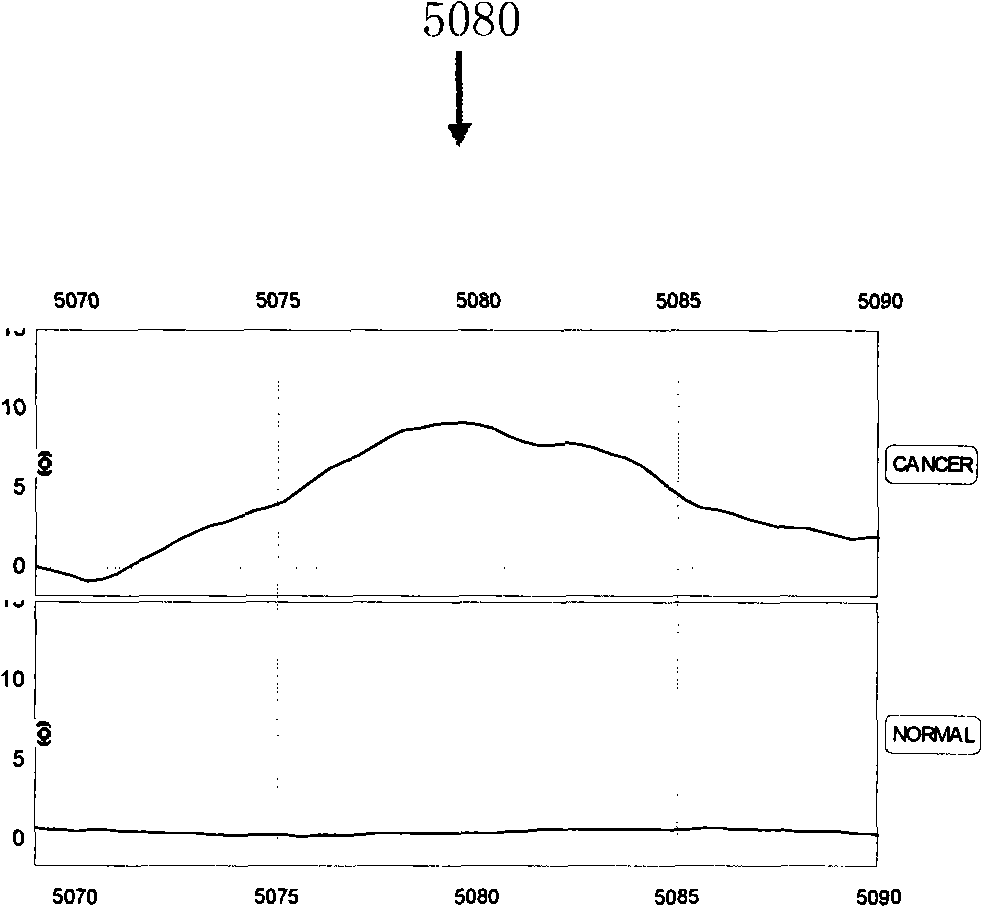
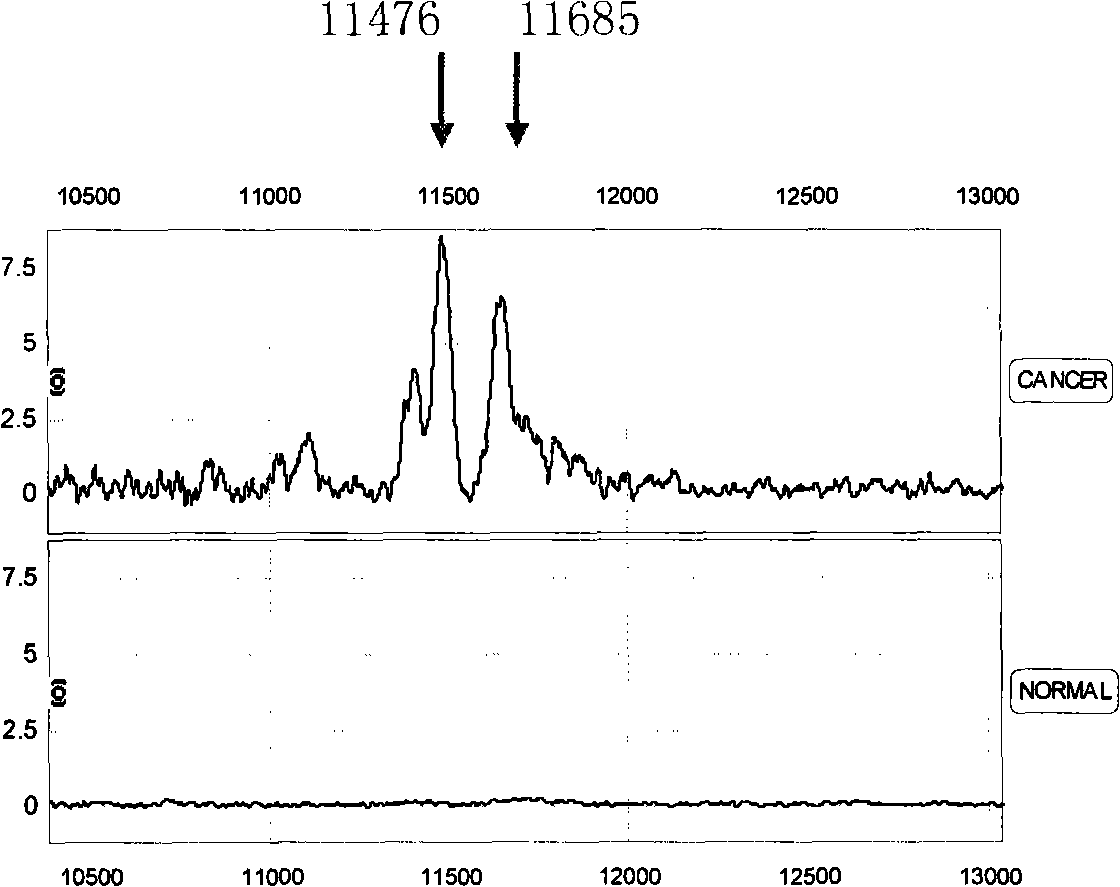
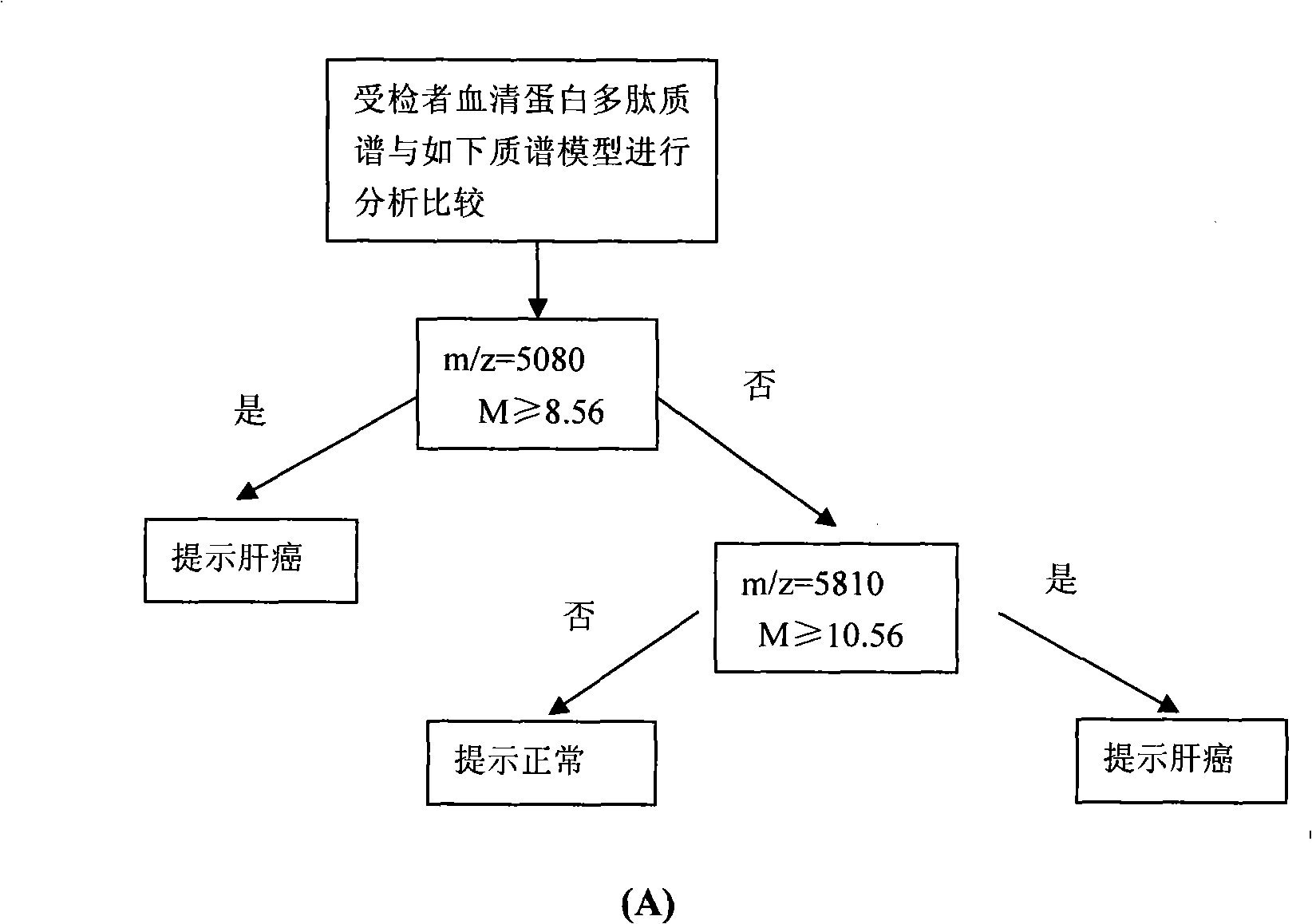
[0081] Because the combination of multiple characteristic proteins can completely separate liver cancer from normal people, so any two or more of the above-mentioned 11 characteristic proteins are selected, and according to the mass-to-charge ratio m / z value of each protein peak and Based on the critical peak value M of the protein, a serum characteristic protein detection mass spectrometry model for pairwise identification of liver cancer patients and normal people, liver cirrhosis caused by hepatitis B, and distant metastasis of liver cancer was established ( Figure 3-5 ), said specific protein mass-to-charge ratio m / z and critical peak value M are respectively m / z=5080, M≥8.56; m / z=5810, M≥10.56; m / z=5335, M≥12.36; m / z=8690, M≤2.87; m / z=4470, M≤20.21; m / z=11685, M≤16.04; m / z=5335, M≥12.36; m / z=8937, M≥10.71; Among them, the mass spectrometry model A for distinguishing liver cancer patients from normal people is dra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com