Reagents, methods, and libraries for bead-based sequencing
A technology of sequence and cutting agent, applied in the field of bead-based sequencing reagents and libraries, can solve the problems of limiting wide application, distinguishing errors of highly similar sequences, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0379] Example 1: Efficient Cleavage and Ligation of Phosphorothioated Oligonucleotides
[0380] This example describes experiments showing efficient ligation and cleavage of extended oligonucleotides containing 3'-S phosphorothioate linkages.
[0381] Materials and methods
[0382] ligation sequencing method
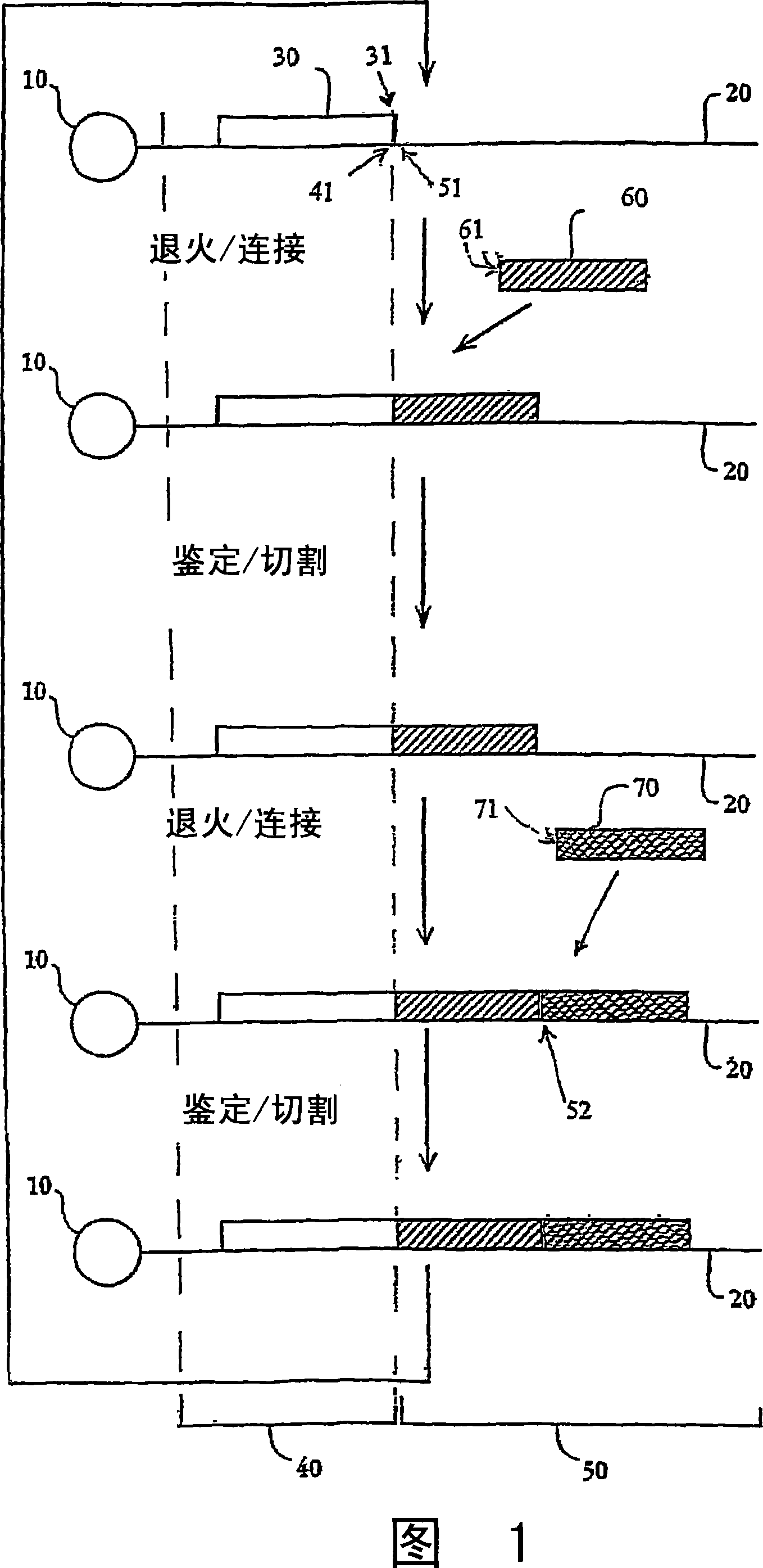
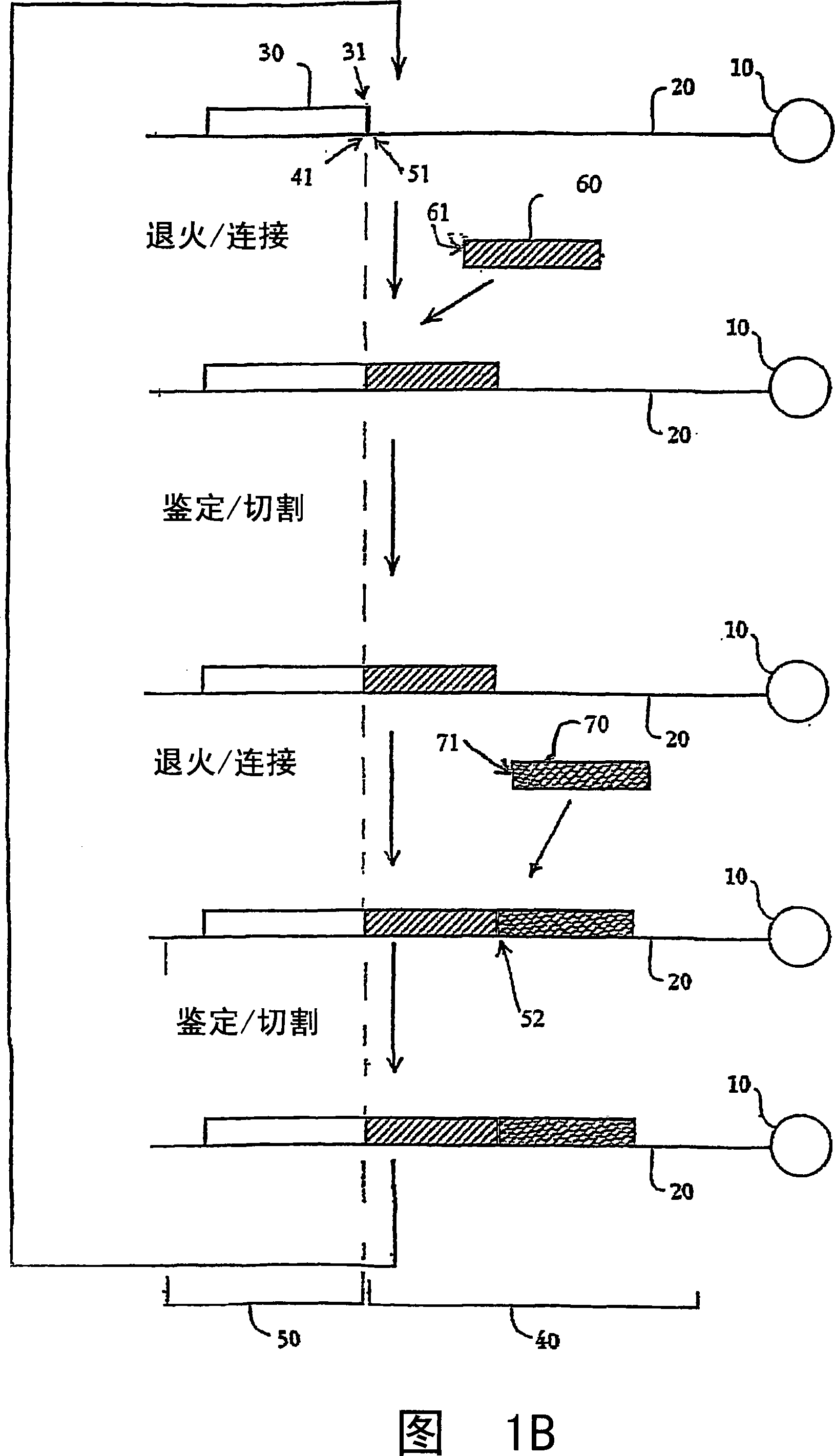
[0383] Template Preparation: To evaluate the potential for sequencing by oligonucleotide ligation and cleavage cycles and to explore the effect of altering certain aspects of the method, two populations of model bead-based templates were prepared. In a preferred implementation, cycles of oligonucleotide ligation and cleavage extend the strand in the 3'→5' direction, as described in the Examples. Therefore, in order to evaluate the ligation efficiency, the 5' end of the pattern template was bound to beads, and the same binding region was designed at the 3' end. One set consisted of short (70 bp) oligonucleotides bound to streptavidin-coated magnetic beads (1 micron)...
Embodiment 2
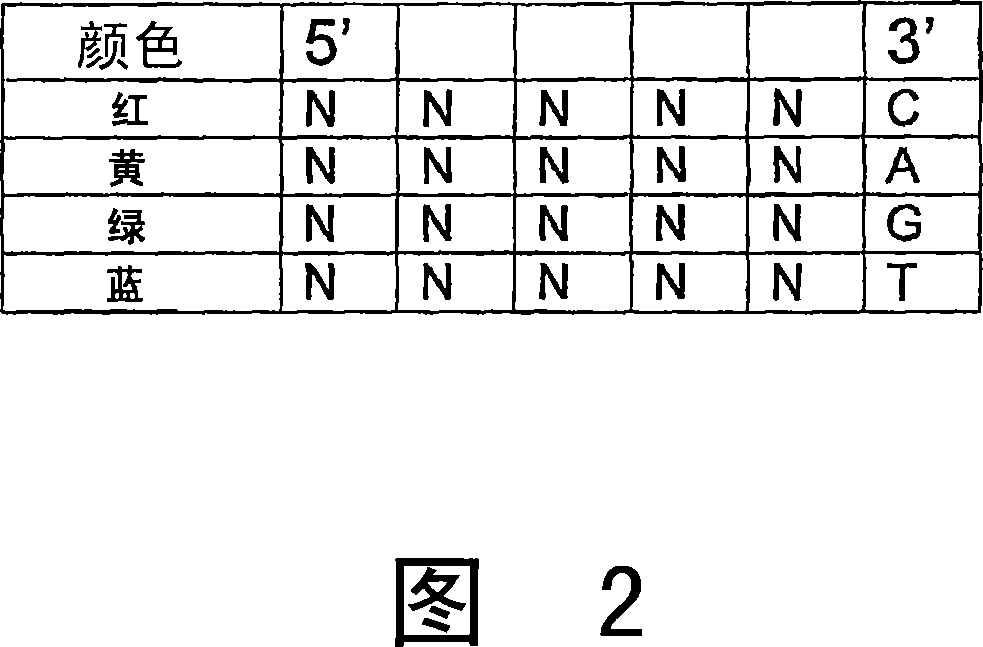
[0397] Example 2: Efficient Cleavage and Ligation of Phosphorothioated Oligonucleotides Containing Nucleotides with Reduced Degeneracy
[0398] However, another consideration for probe length is the fidelity of the extended oligo and its impact on subsequent ligation efficiency. The fidelity of T4 DNA ligase has been shown to decrease rapidly after the 5th base after the junction (Luo et al., Nucleic Acids Res., 24:3071-3078 and 3079-3085, 1996). If a mismatch is introduced on the 5' side of the newly ligated junction, ligation efficiency can be reduced by depletion, however, without a phase shift or increase in background signal (a major hurdle encountered in polymerase-based sequencing by synthetic approaches). ).
[0399] Preferably, the probe set should be able to hybridize to any DNA sequence in order to resequence uncharacterized DNA. However, the complexity of labeled probe sets increases exponentially with the length and number of 4-fold degenerate bases. Furthermor...
Embodiment 3
[0404] Example 3: Fidelity of Probe Ligation
[0405] Bacterial NAD-dependent ligases such as Taq DNA ligase have been reported to have high sequence fidelity at the ligation, with essentially no nick-closing activity for mismatches on the 3' side but some tolerance for mismatches on the 5' side (Luo et al., Nucleic Acids Res., 24:3071-3078 and 3079-3085, 1996). On the other hand, T4 DNA ligase has been reported to be slightly less stringent, allowing mismatches at the 3'- and 5'-sides of the junction. Therefore, it was of interest to evaluate the fidelity of probe ligation with T4 DNA ligase compared to Taq DNA ligase in our system.
[0406] Using standard ABI sequencing techniques, we developed two methods to assess the sequence fidelity of ligated oligonucleotides. The first approach was designed to clone and sequence the ligation products. In this method, ligated extension products are ligated to adapter sequences, cloned and transformed into bacteria. Individual colon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com