Method of targeting specific cell populations using cell-binding agent maytansinoid conjugates linked via a non-cleavable linker, said conjugates, and methods of making said conjugates
A technology of maytansinoids and alkaloids, which can be applied to medical preparations with non-active ingredients, medical preparations containing active ingredients, and pharmaceutical formulas, and can solve insurmountable problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1A
[0254] Preparation of huC242-SMCC-DM1 conjugate
[0255] a. Preparation and determination of huC242 antibody
[0256] [185] with an extinction coefficient of 1.48 (mg / mL) at 280nm -1 and a molecular weight of 147,000 g / mole, the antibody concentration was measured.
[0257] b. Preparation and determination of SMCC stock solution
[0258] [186] A 20 mM solution of SMCC (6.69 mg / mL) was prepared in dimethylsulfoxide. The solution was diluted 1 / 40 in Assay Buffer, and the absorbance of the sample was measured at 302 nm. With extinction coefficient 602M -1 cm -1 Calculate the concentration of the stock solution.
[0259] c. Preparation and determination of DM1 stock solution
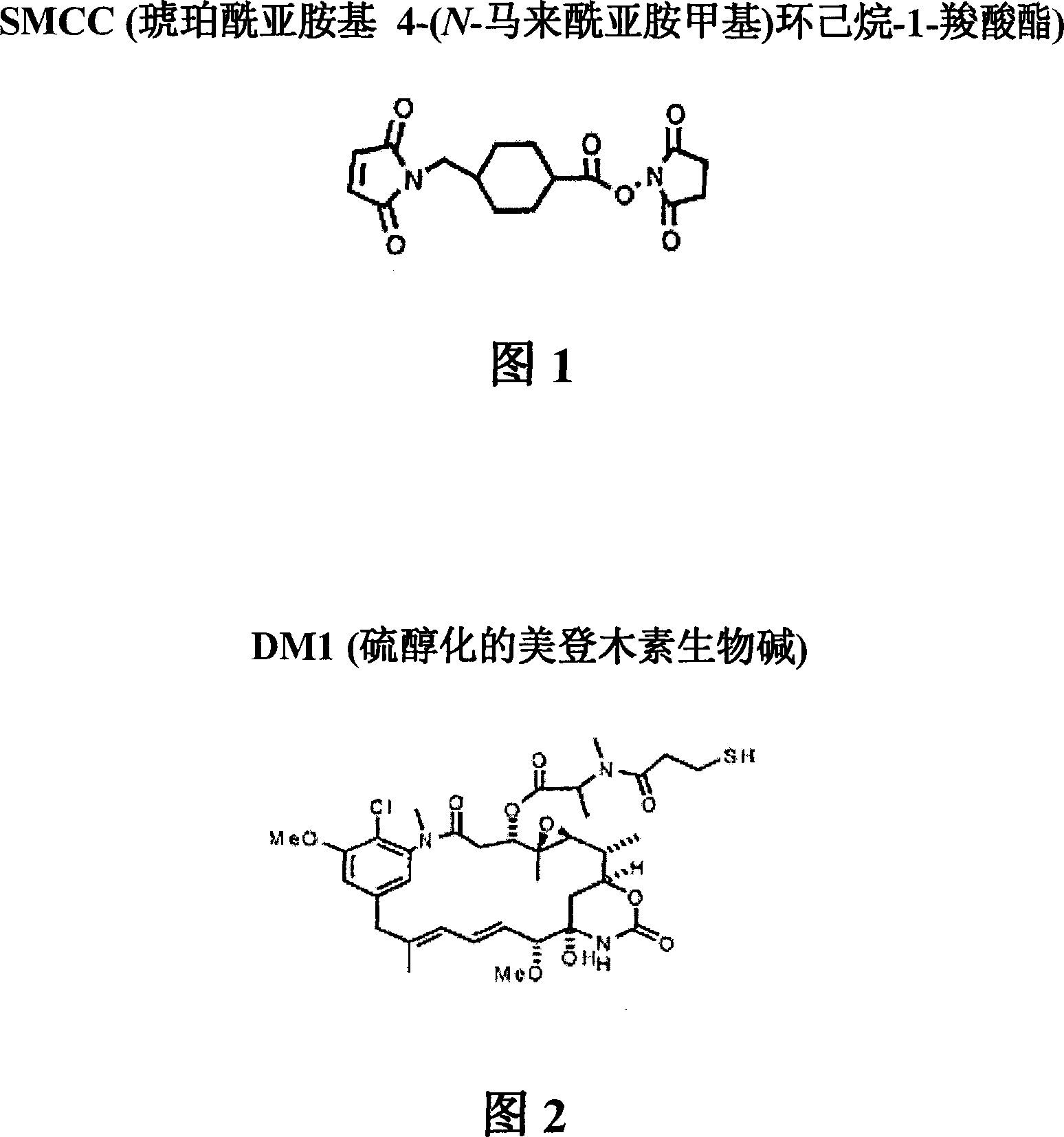
[0260] [187] A 10 mM solution of DM1 (free thiol form) (7.37 mg / mL) was prepared in dimethylacetamide (DMA) (Figure 2). The absorbance of a dilution of the stock solution in ethanol (EtOH) was measured at 280 nm. With an extinction coefficient of 5700M at 280nm -1 Calculate the concentration of sto...
Embodiment 1B
[0272] In vitro detection of huC242-SMCC-DM1
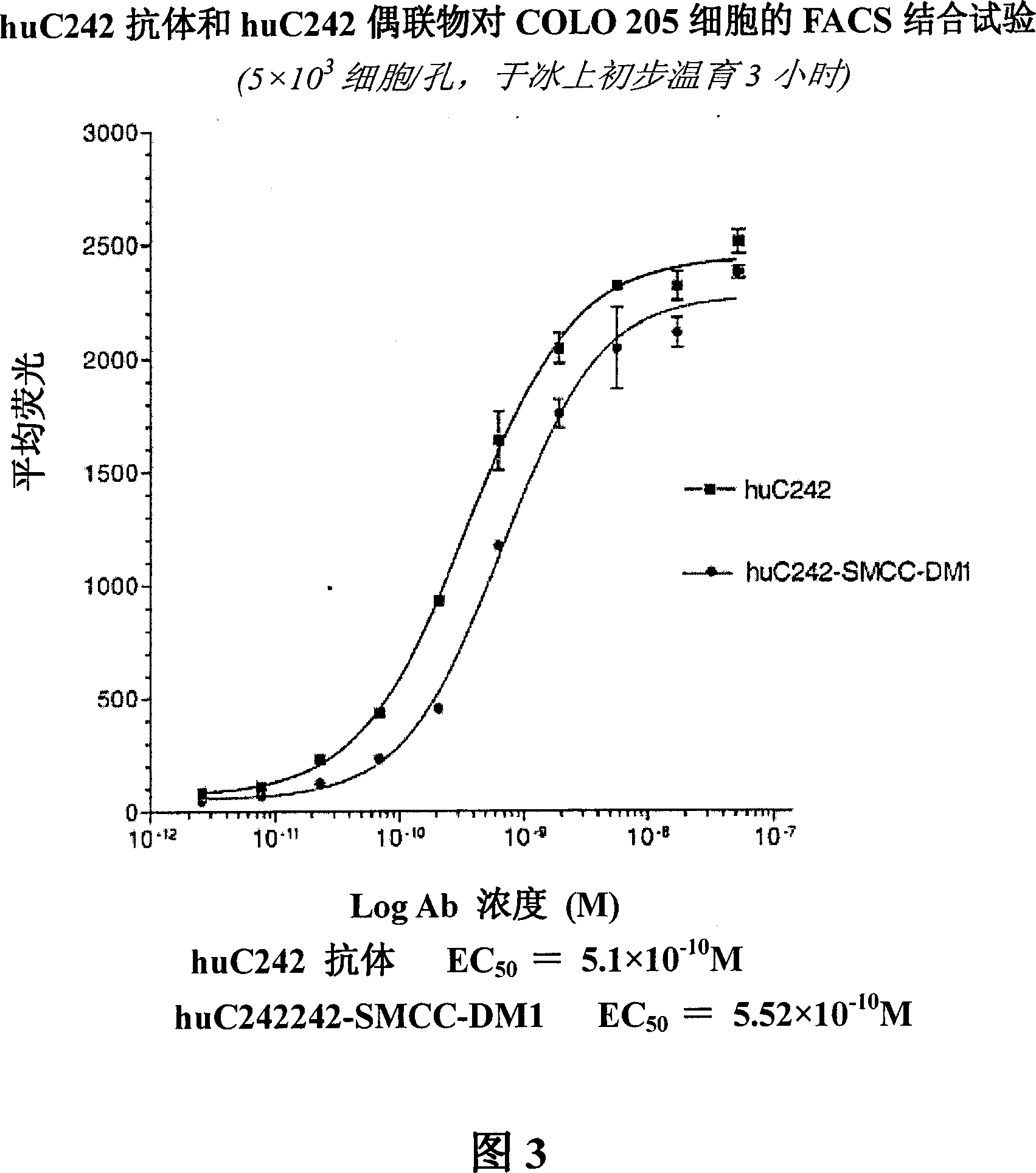
[0273] a. Binding
[0274] [192] The binding affinities of the huC242 antibody and huC242-SMCC-DM1 were compared using an indirect method on COLO205 cells. The results are shown in Figure 3. Naked antibody (naked antibody) at 5.1×10 -1 The KD binding of M, the conjugate is 5.52×10 -10 KD binding of M. Therefore, linking to DM1 does not appear to alter the binding affinity of huC242.
[0275] b. Cytotoxicity and Specificity
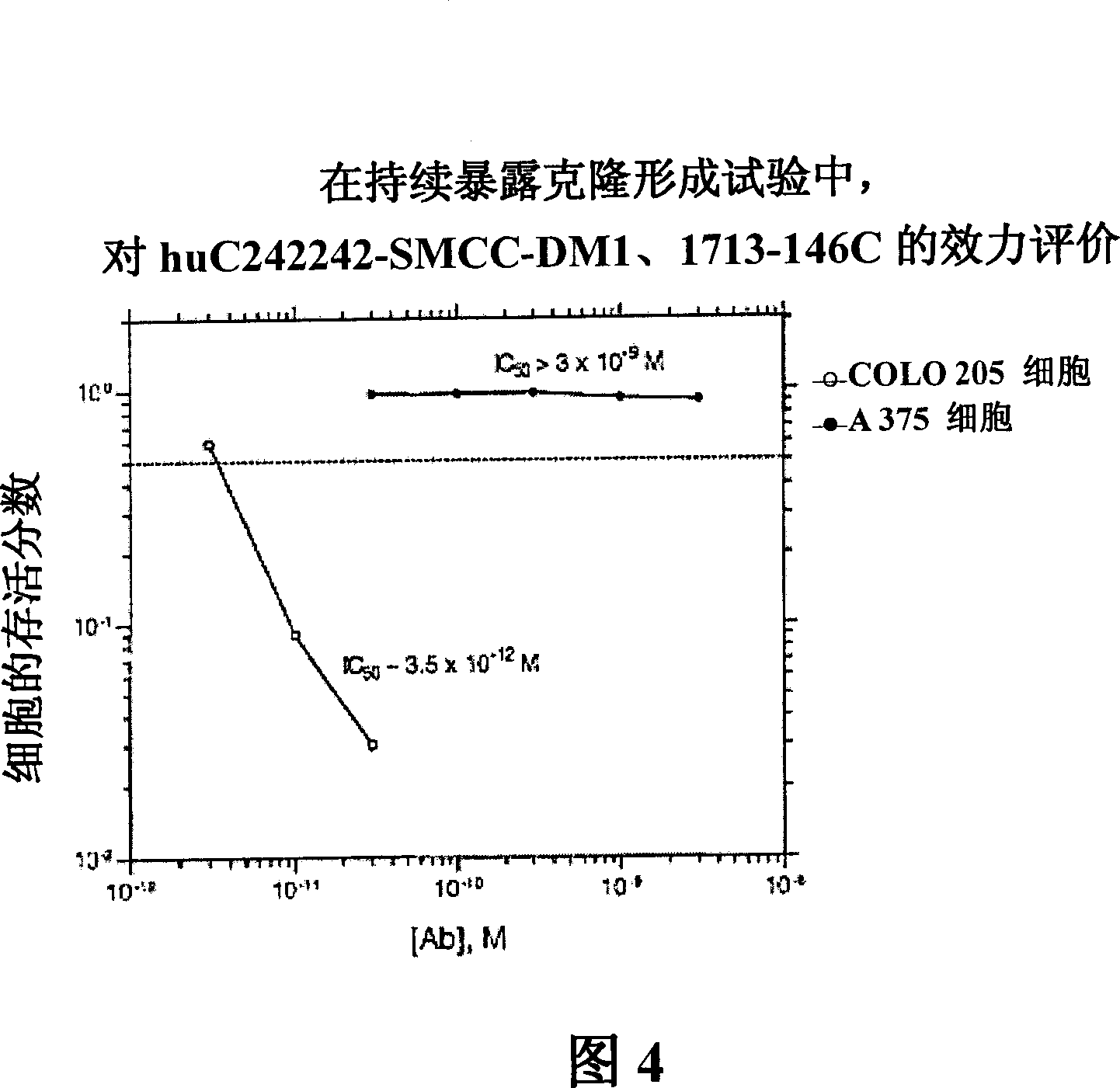
[0276][193] The in vitro cytotoxicity and specificity of the huC242-SMCC-DM1 conjugate was evaluated using a continuous exposure clonogenic assay. The results are shown in Figure 4. huC242-SMCC-DM1 was effective in destroying antigen-positive SKBR3 cells (IC 50 =3.5×10 -12 M). Specificity is determined by comparing the IC of target SKBR3 cells 50 value and the IC of the antigen-negative cell line A375 50 Values are shown where the IC of the conjugate 50 Value greater than 3.0×10 -9 M.
[0277]...
Embodiment 2A
[0282] Preparation of trastuzumab-SMCC-DM1 conjugate
[0283] [196] The antibody trastuzumab was obtained from Genentech for linking with the non-cleavable heterobifunctional cross-linkers SMCC and DM1. Antibodies were buffer exchanged from 50 mM potassium phosphate / 2 mM EDTA, pH 6.0 into 50 mM potassium phosphate / 50 mM sodium chloride / 2 mM EDTA, pH 6.5 (buffer A). The antibody was then reacted with a 7.5-fold molar excess of the SMCC linker and purified on Sephadex G25 resin prior to ligation of the antibody to DM1. The final conjugate was purified again with Sephadex G25 resin. The resulting conjugate contained 3.1 moles of DM1 per mole of antibody.
[0284] a. Preparation and Assay of Antibody Trastuzumab
[0285] [197] Trastuzumab dissolved in 50 mM potassium phosphate / 2 mM EDTA buffer, pH 6.0, was passed through a Sephadex G25 column equilibrated with buffer A and eluted as buffer A. All buffers in this assay were tested to be free of endotoxin using the chromogenic L...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extinction coefficient | aaaaa | aaaaa |
| Extinction coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com