Method of producing mutation procarboxypeptidase B and mutation carboxypeptidase B
A technology of pro-carboxypeptidase and carboxypeptidase, which is applied in the field of molecular biology and can solve the problem that the mutation of CPB by site-directed mutagenesis technology is not reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1: Construction of mCPB / mPCPB expression plasmid pET-mCPB / pET-mPCPB
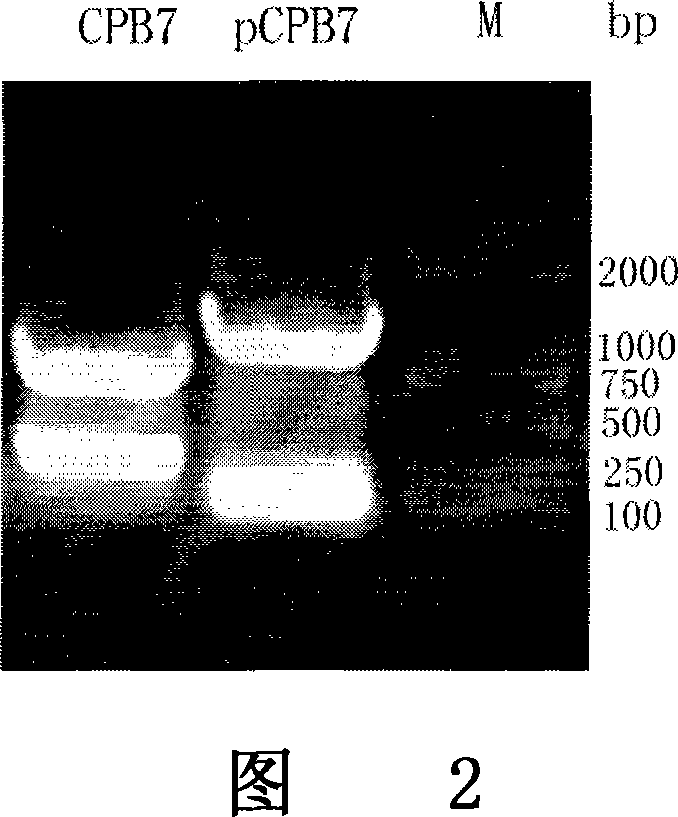
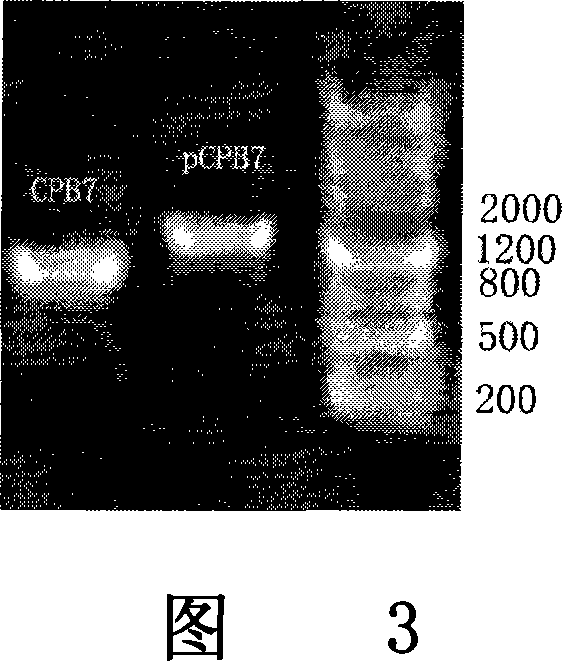
[0077] I. The mPCPB gene is derived from the constructed plasmid pET-21a-pCPB containing the procarboxypeptidase B gene, which is obtained from the pancreas of Wistar rats by RT-PCR and obtained after site-directed mutation. (As shown in Figure 1).
[0078] PCR amplification mCPB / mPCPB gene, the DNA nucleotide sequence of three primers used in the cloning: be respectively the 5' end primer of PCPB and the 3' end primer (primer 1 (SEQ ID NO: 3) and primer 3 (SEQ ID NO: 3) IDNO: 5)); CPB's 5' end primer and 3' end primer (primer 2 (SEQ ID NO: 4) and primer 3 (SEQ ID NO: 5)), and another antisense primer was used to introduce Hind III restriction site Stop codon after dot (SEQ ID NO: 6). Nco I / Hind III double restriction site, Nco I / Hind III double restriction site and stop codon were introduced respectively.
[0079] Primer 1: pCPB: sense primer, 5'-GCG GGA TCC CAT GCT TCC GAGGAG CAC TTT GAT ...
Embodiment 2
[0095] Example 2: Screening of recombinant mutant mPCPB clones by inducing expression of mutants by SDS-PAGE
[0096] I. Medium
[0097] After transformation, Escherichia coli BL21(DE3) carrying the recombinant plasmid pET-mPCPB can grow on solid LB medium containing ampicillin (100 μg / ml).
[0098] II. Inoculum
[0099] Pick a single colony and put it in 5ml LB (containing 100μg / ml ampicillin) liquid medium, shake the bacteria overnight at 37°C; inoculate overnight bacteria at 1% in 250ml LB ampicillin-resistant culture medium, shake the bacteria at 37°C The OD at 600nm is 0.3-0.5, induced by 0.5mM IPTG, and adjusted to the corresponding temperature. After 3-5 hours after induction, the bacteria are collected by centrifugation.
[0100] III. Expression Analysis
[0101] Centrifuge the cells before and after induction, discard the supernatant, add electrophoresis sample buffer to the precipitated cells, boil for 10 min, centrifuge, and analyze the supernatant by denaturing ...
Embodiment 3
[0102] Example 3: Induced expression of mutants SDS-PAGE for screening of recombinant mutant mCPB clones
[0103] The screening method is the same as in Example 2. The results are shown in Figure 7.
[0104] Compared with before induction, the corresponding protein band (35KD) was expressed after being induced by 0.5mM IPTG containing the recombinant plasmid. The results showed that strains 1 and 3 were positive clones, and strain 2 was a negative clone.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com