Methods and compositions for manipulating translation of protein isoforms from alternative initiation start sites
a protein isoform and initiation start technology, applied in the field of antisense oligonucleotides and compositions containing antisense oligonucleotides, can solve the problems of reducing the translation efficiency of p53, affecting the expression quality and quantitative differences, and directing much effort to reconstitute or reactivate p53 expression, etc., to achieve the effect of enhancing the sensitivity of tumor cells to radiation therapy, enhancing the translation of a second
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
pMOS Block P53 Expression and Generate P53 Isoforms
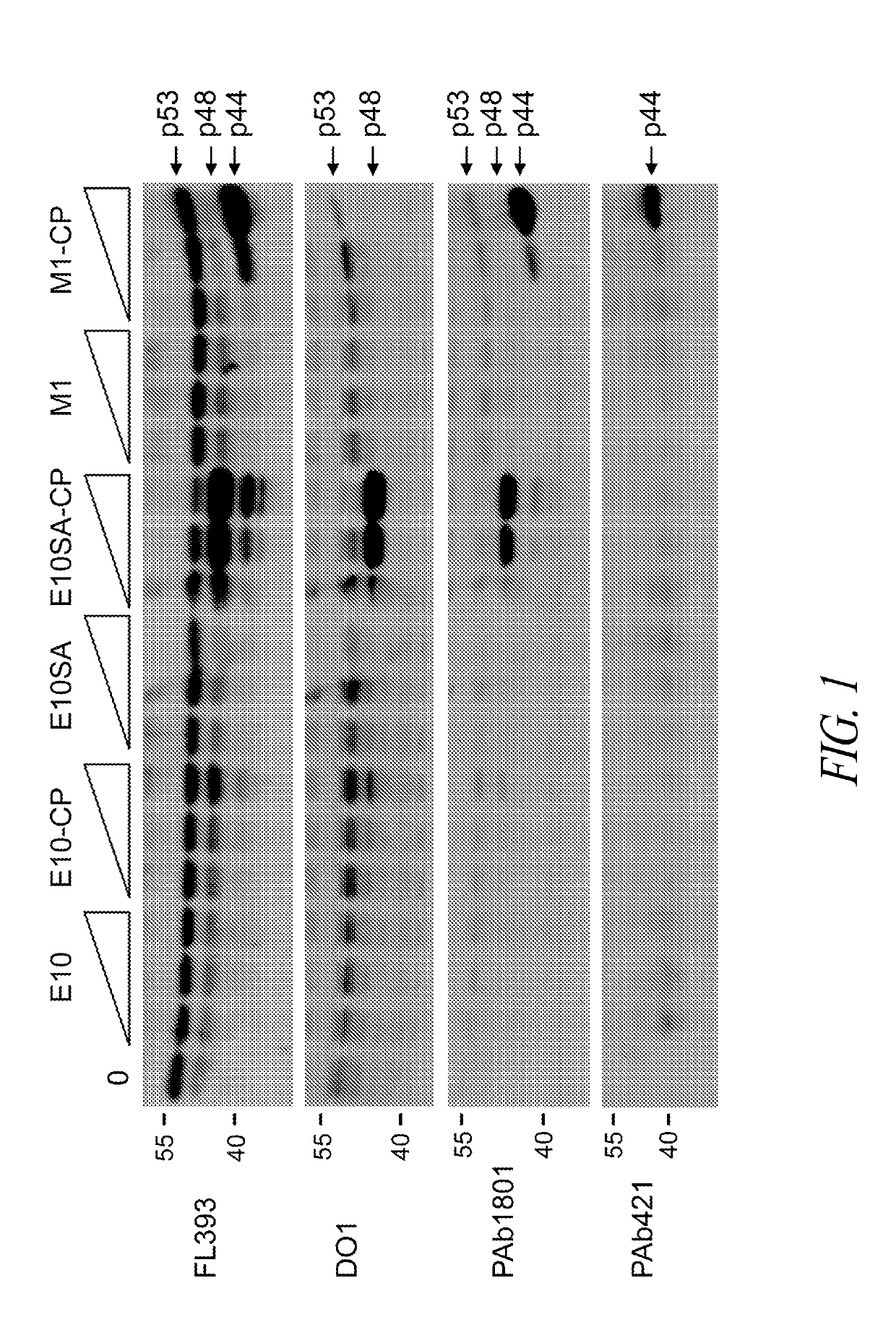
[0388]To investigate the effectiveness of PMOs in blocking p53 expression, wild-type p53 expressing breast cancer MCF7 cells were treated with PMOs and peptide-conjugated PMOs targeting the translation initiation site of p53 mRNA (M1), the splice acceptor site of exon 10 (E10SA) and the coding region within exon 10 (E10) (see Table A below for PMOs used in all experiments described herein).
TABLE Ap53 Targeting SequencesSEQPMOp53 RegionTargeting Sequence 5′-3′ID NO:M1AUG startGCGGCTCCTCCATGGCAGTGAC 3codonM40Met 40 AUGCATCAAATCATCCATTGCTTGG 4codonE2SDExon 2AGTTTCCATAGGTCTGAAAA 5E4SAIntron 3 / GIIIACTGTAGATGIGTGAA 6Exon 4E6SAExon 6CGGATAAGATGCTGAGGAGG 7E7SAExon 7GTTGTAGTGGATGGTGGTA 8E7SDExon 7CTGGAGTCTTCCAGTGTGAT 9E9SDExon 9AAGIGTGAAATATTCTCCATC10E10SAIntron 9 / GCGCTCACGCCCACGGATC11Exon 10E10Exon 10CCCTGCTCCCCCCTGGCTCC12(Internal)ControlScrambledTGCCATCAACATATCTTGATCG13ControlSD, slice donorSA, splice acceptorI, inosine residue
[0389]The p...
example 2
pMOS Block P53 Expression and P21 Induction after DNA Damage
[0394]To investigate the expression and function of p44 (ΔN-p53) and p48 (p53β / γ) isoforms in a broader range of cells under different stress conditions that activate p53, human H460 lung cancer cells were treated with doxorubicin, human HCT116 colon cancer cells were treated with 5-FU, and human OCI / AML-3 and OCI / AML-4 leukemia cells were treated with γ-radiation. All of these human cancer cell lines express wild-type p53 and respond to DNA damage by increasing p53 protein levels and activating expression of the p53-target gene, p21 (see FIG. 2).
[0395]As shown in FIG. 2, pre-treatment for 2 hours with peptide-conjugated M1 or E10SA prior to DNA damage led to the production of ΔN-p53 and p53β / γ, respectively, as seen previously in MCF7 cells. The levels of these stable p53 isoforms did not increase further after DNA damage. Both M1 and E10SA interfered with the accumulation of full-length p53 and with the induction of p21 a...
example 3
Evaluation of pMOS that Target P53 mRNA Splicing
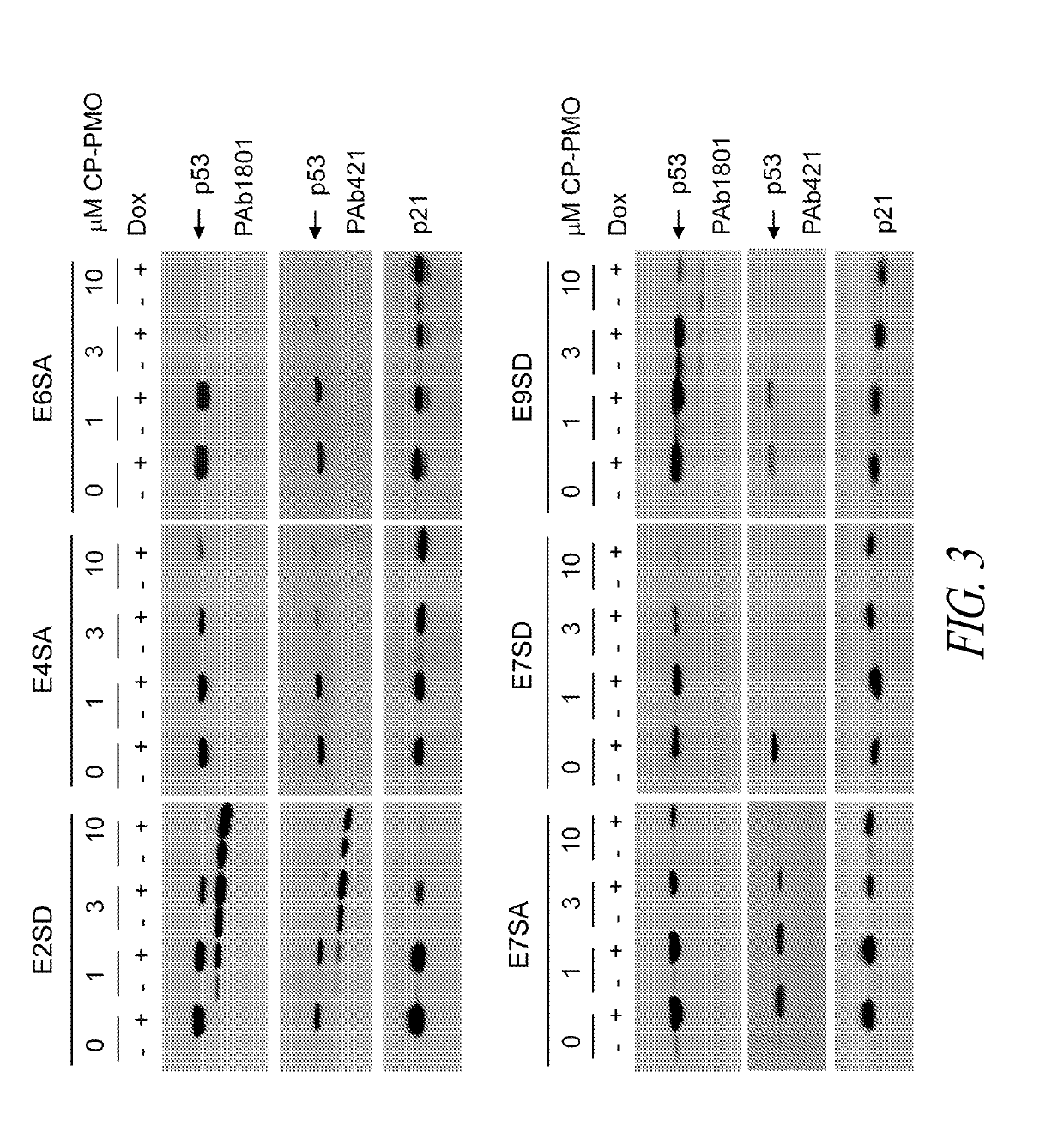
[0397]In the following experiments, a panel peptide-conjugated PMOs was tested that target splice sites in exons 2, 4, 6, 7 and 9 of the human p53 gene (see Table A, in Example 1 above). With the exception of E2SD that targets the splice donor site of exon 2, the remaining PMOs exhibited variable effectiveness in suppressing p53 protein expression in H460 cells, revealed no new isoforms, and did not block expression of p21 in response to doxorubicin (see FIG. 3).
[0398]As shown in FIG. 3, E2SD produced a 44 kDa isoform that resembled ΔN-p53 and prevented the induction of p21 in response to doxorubicin treatment. This alternatively spliced transcript contains an intron 2-derived in-frame termination codon that arrests translation prematurely and was predicted to produce ΔN-p53 from codon 40. These findings suggest that E2SD and M1 produce identical truncated ΔN-p53 proteins through two different mechanisms: alternative splicing of exon 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com