Protease stable cell wall lysing enzymes
a stable, cell wall technology, applied in the direction of instruments, antibacterial agents, peptide/protein ingredients, etc., can solve the problems of only being used in limited quantities by protease inhibitors for stabilizing and affecting the stability of the etc., to achieve the effect of stable cell wall lysing enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Protease Digest of PlyPitti26
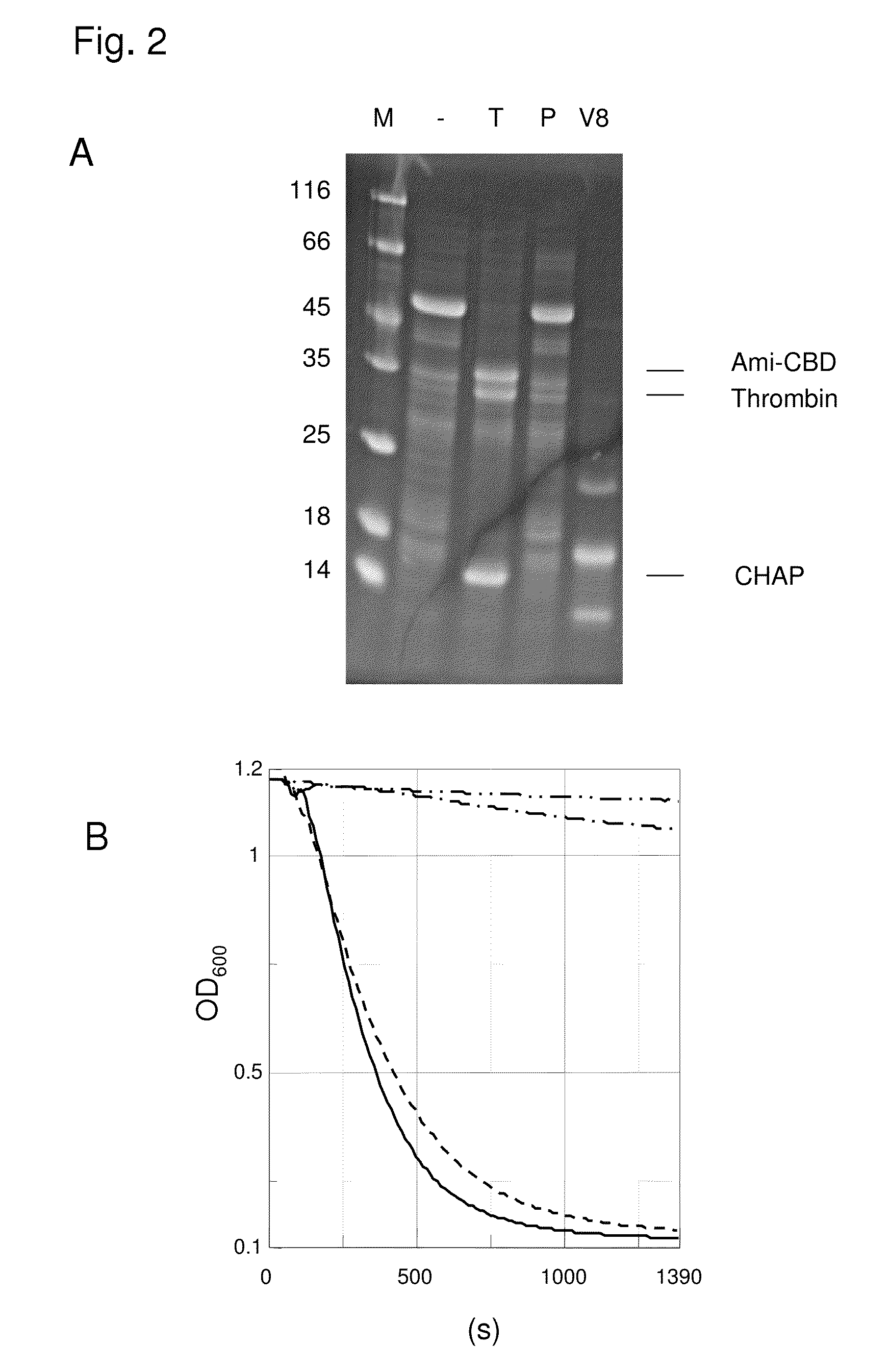
[0069]It was performed a digest of PlyPitti26 with the proteases thrombin and V8. Each 90 μl protein solution (protein concentration 1.35 mg / ml) were tampered with 10 μl thrombin and in control samples only tempered with plasmin. For the V8 protease digest 99 μl protein solution and 1 protease solution were used. The concentration of the protease stock solutions was each 500 μg / ml. The samples were incubated in a 20 mM Tris / HCl pH 7.5, 10 mM DTE, 0.1 mM ZnSO4 Buffer over night at 25° C. A part of the samples was tempered with SDS gel-application buffer and was analysed on SDS polyacrylamide gels after boiling.
[0070]It was found that PlyPitti26 is degraded completely by thrombin, wherein two relative large protein fragments emerged, of which the one contained the CHAP-domain and the other the amidase domain and the CBD. In a digest with V8 protease several smaller, but also defined protein fragments emerged. In a control digest with plasmin no fragmentati...
example 2
Activity Test of Untreated PlyPitti26 and PlyPitti26 Treated with Proteases
[0071]In further experiments the samples were tested in the liquid lysis test on the lysis activity towards Staphylococcus cells. The liquid lysis test is an activity test for cell wall lysing enzymes, in which the bacterial cell lysis was measured in real time via light scattering in the photometer. The absorption at a wave length of 600 nm is thereby a measure for the number of existing bacteria cells. If the bacteria lyse the sample solution is getting clear by degrees and the measured absorption decreases in the ideal case down to a value of zero. Staphylococcus bacteria were cultivated in BHI medium to an OD600 of about 0.8. The not heat activated cells were harvested and resuspended to an OD600 of about 1 in TBST buffer (20 mM Tris / HCl pH 7.5, 60 mM NaCl, 0.1% Tween, 2 mM CaCl2). 990 μl cell suspension was tempered each with 10 μl enzyme solution (enzyme concentration in the test 10 μg / ml) and the decre...
example 3
Stabilization of CHAP-AmiPitti26-CBDUSA
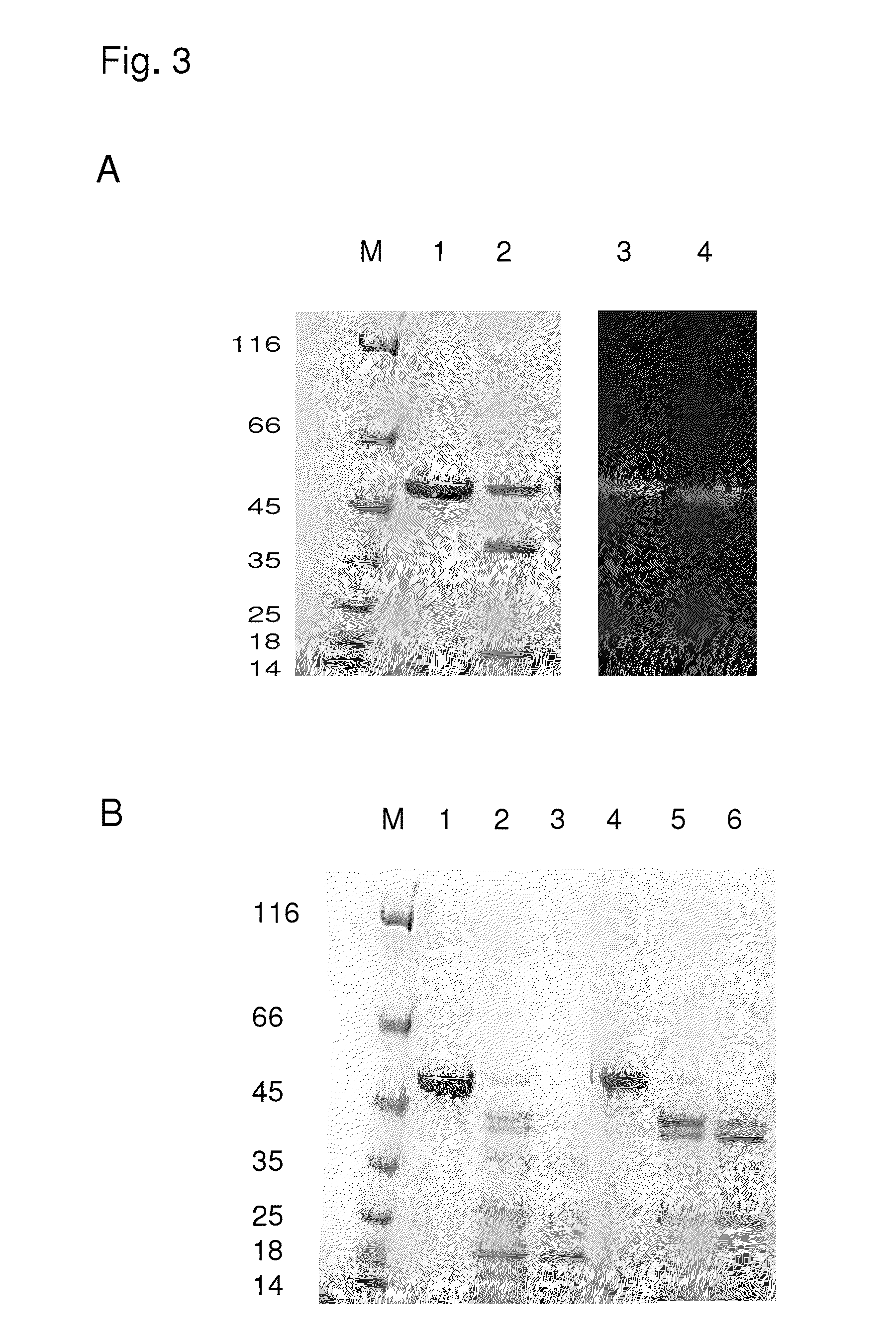
[0073]A synthetically produced endolysin variant active against staphylococci, which consists of the enzymatically active domains CHAP and Ami of the endolysins of PlyPitti26 and of the cell wall binding domain (CBD) of the endolysin of the prophage Φ SA2USA deriving from the genome of the MRSA strain USA300 (Diep et al., The Lancet, 2006, 367, 731-739; database accession number NC—007793) is described as CHAP-AmiPitti26-CBDUSA.
[0074]To stabilize this protein against thrombin digest and digest by V8 protease, both the thrombin cleavage site and V8 cleavage sites lying in the linker between the CHAP- and amidase domains were modified by amino acid substitutions by means of site-directed mutagenesis. The single, double, triple and quadruple mutations performed for that purpose are summarized in table 3. For all mutations given in table 3, endolysin activity against staphylococci was still detected in the plate lysis test, as described below after...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Biological properties | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com