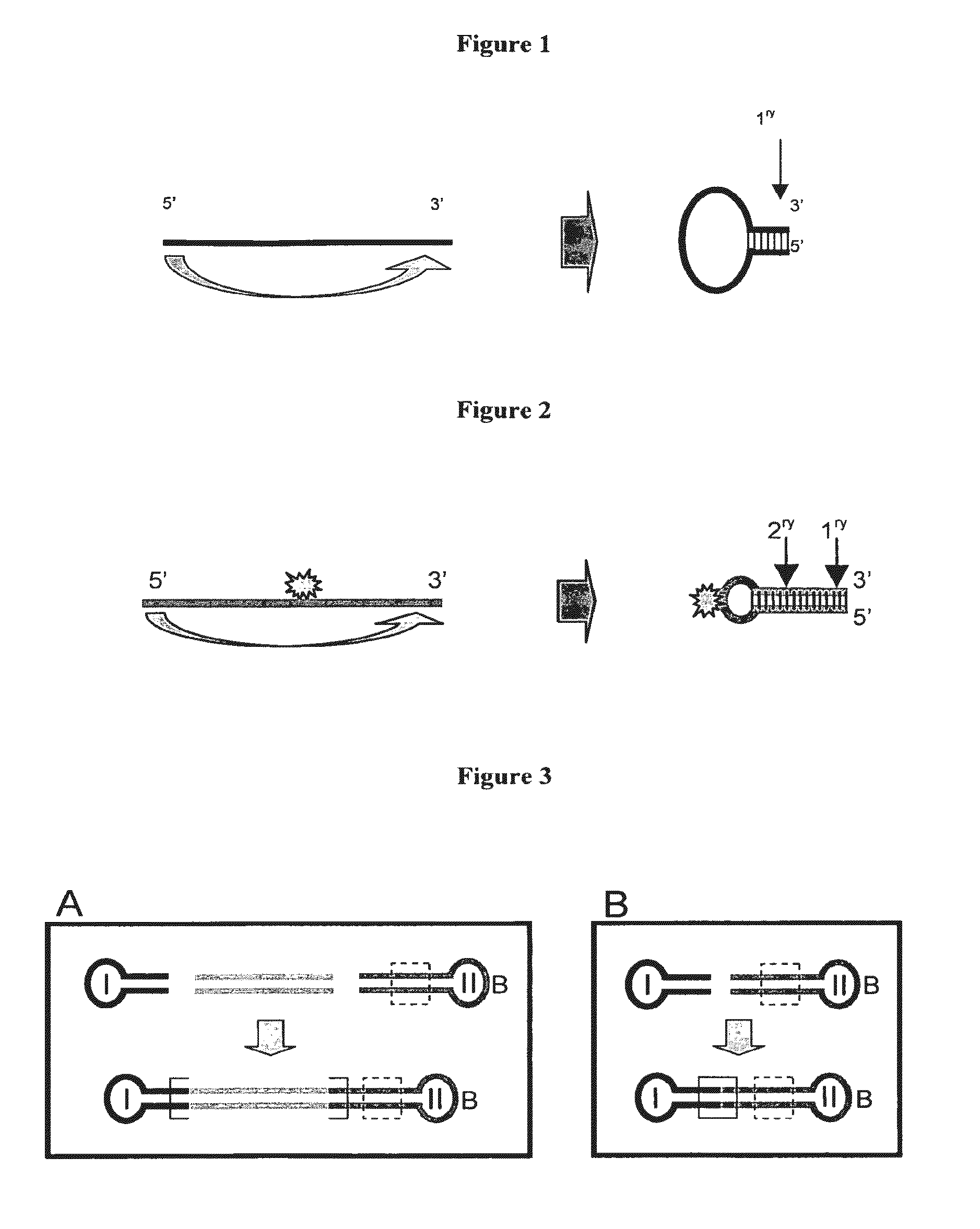
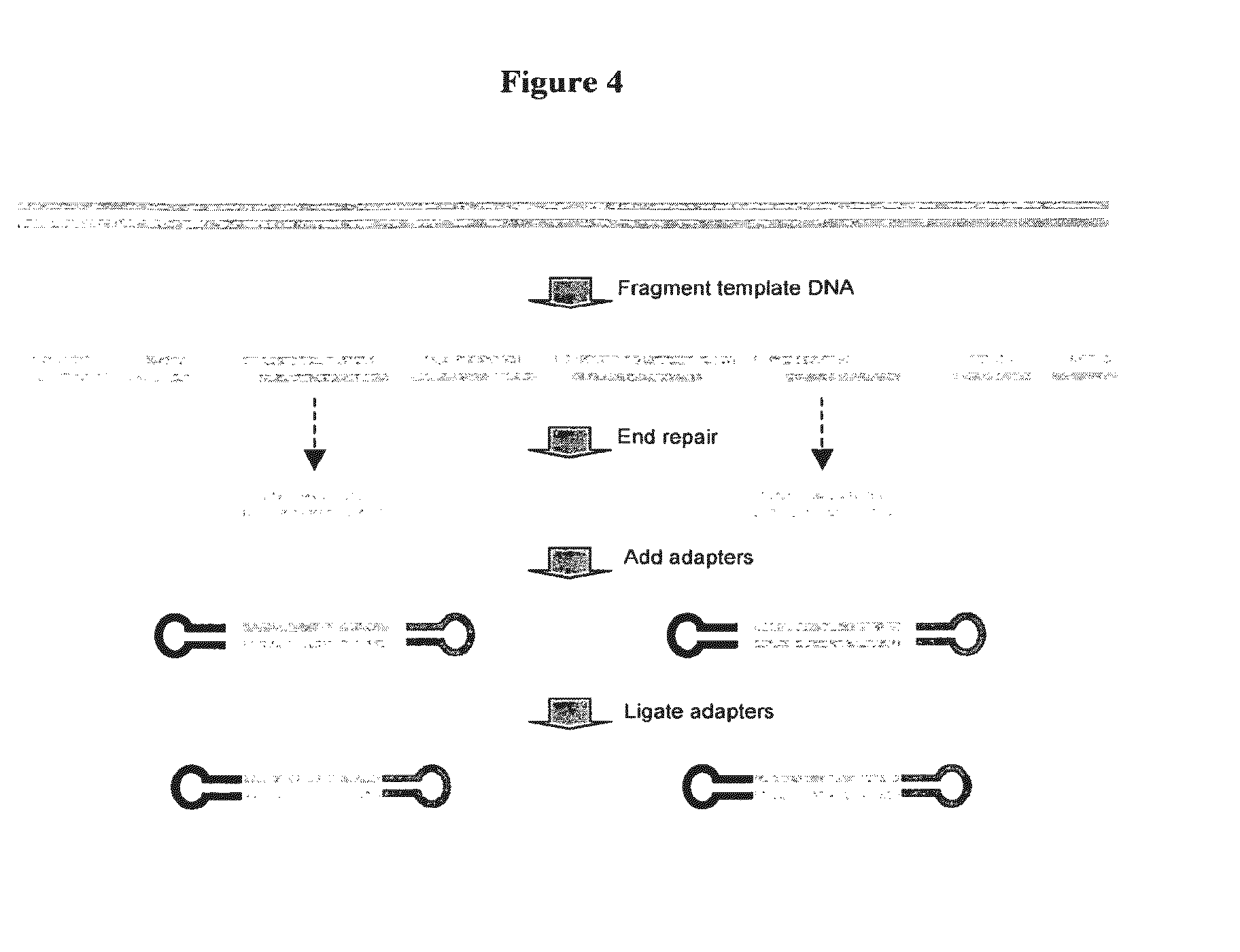
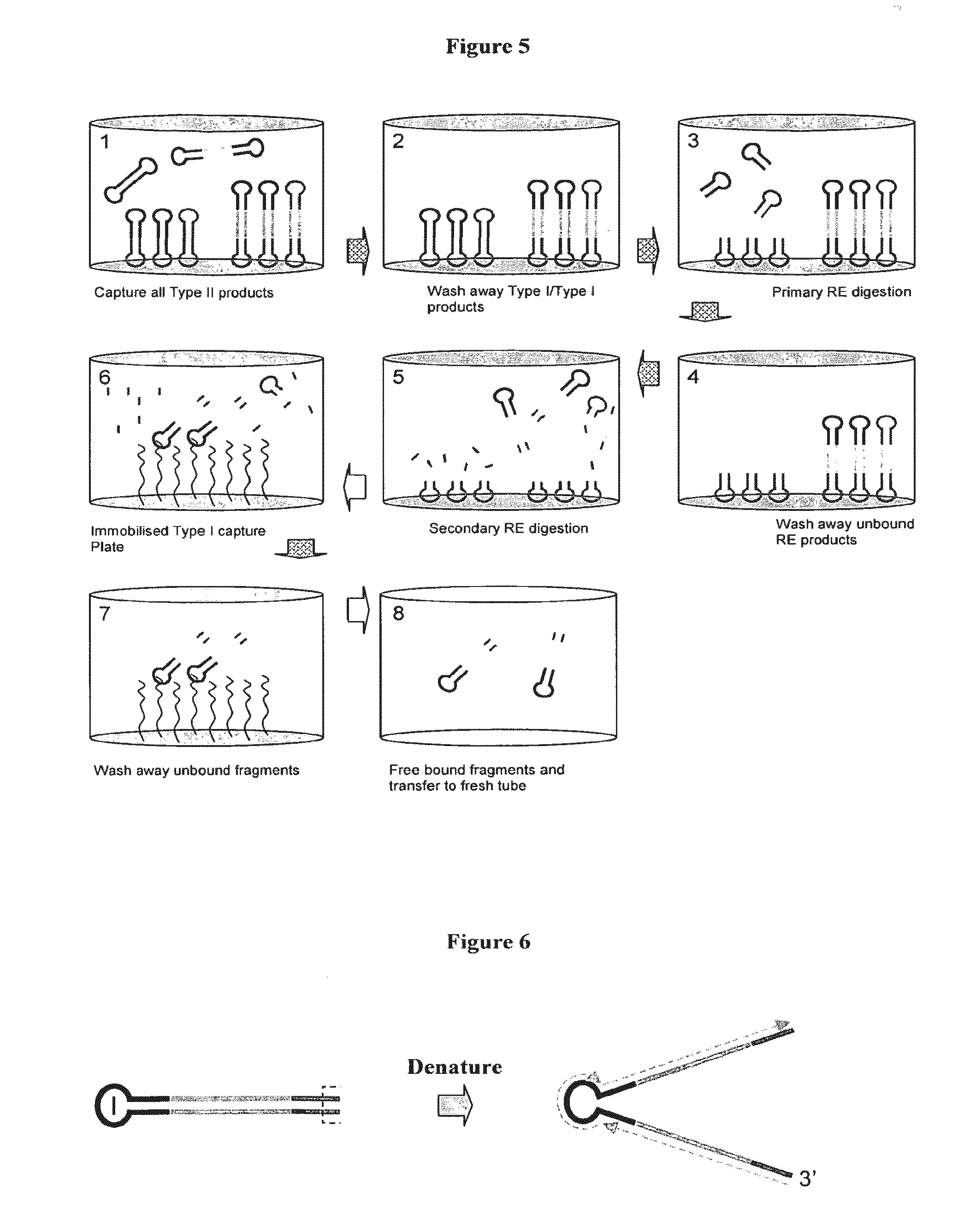
Adaptors for nucleic acid constructs in transmembrane sequencing
a technology of adaptors and nucleic acids, applied in the field of adaptors for sequencing nucleic acids, can solve the problems of slow and expensive existing technologies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0067]It is to be understood that different applications of the disclosed products and methods may be tailored to the specific needs in the art. It is also to be understood that the terminology used herein is for the purpose of describing particular embodiments of the invention only, and is not intended to be limiting.
[0068]In addition as used in this specification and the appended claims, the singular forms “a”, “an”, and “the” include plural referents unless the content clearly dictates otherwise. Thus, for example, reference to “a construct” includes “constructs”, reference to “a transmembrane pore” includes two or more such pores, reference to “a molecular adaptor” includes two or more such adaptors, and the like.
[0069]All publications, patents and patent applications cited herein, whether supra or infra, are hereby incorporated by reference in their entirety.
Adaptor
[0070]The invention provides adaptors for sequencing nucleic acids. The adaptors comprise a region of double stran...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Cell angle | aaaaa | aaaaa |
| Cell angle | aaaaa | aaaaa |
| Acidity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com