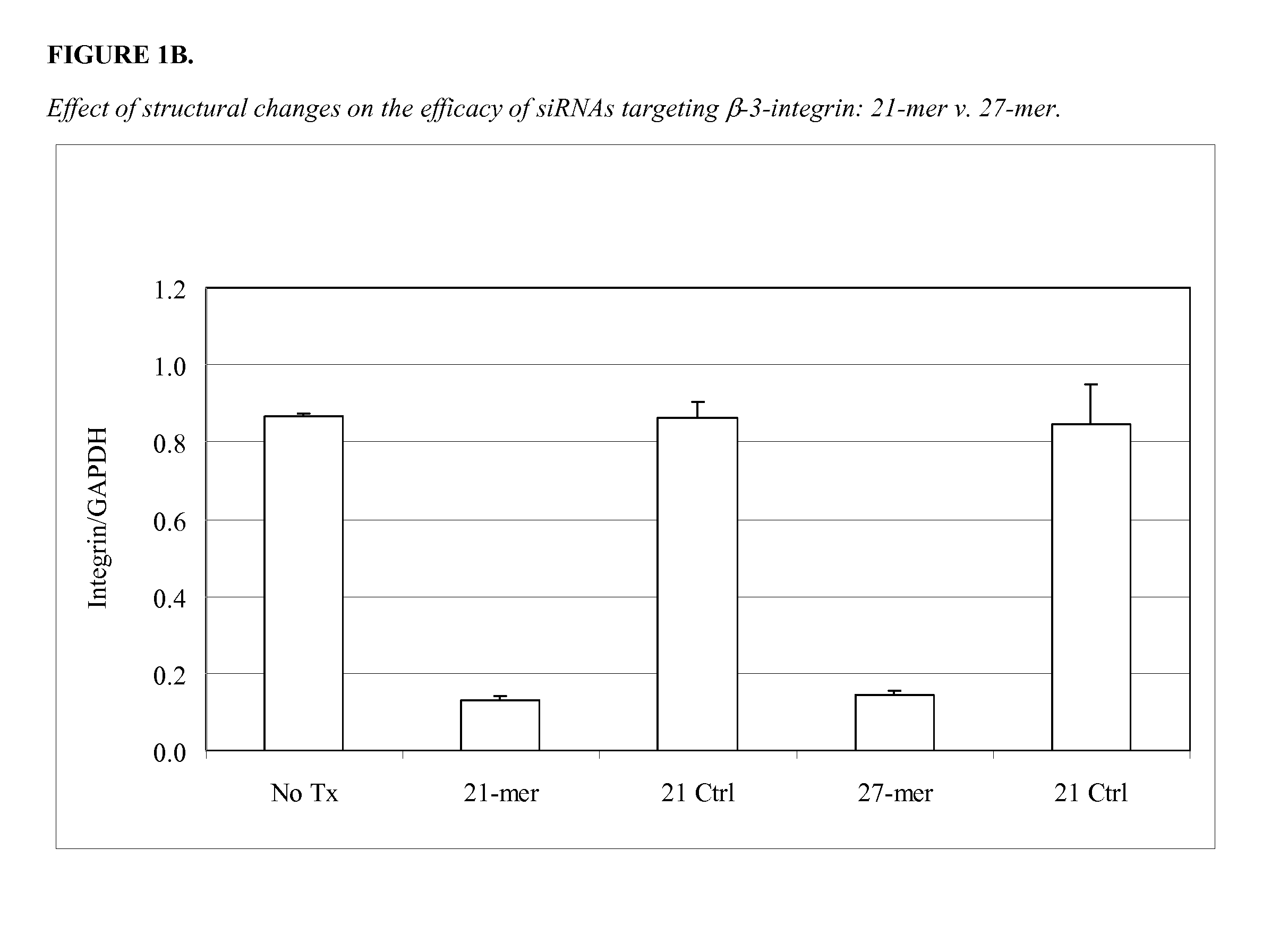Double-Stranded Oligonucleotides
a technology of oligonucleotides and oligonucleotide molecules, which is applied in the direction of transferases, drug compositions, immunological disorders, etc., can solve the problems of difficult to achieve efficient inhibition of gene expression (including protein synthesis) using such compositions, and often subject to nuclease degradation, so as to improve the antisense sequence of prior art and inhibit gene function
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Oligonucleotide Compositions Comprising Chimeric Antisense Sequences
[0352]A gapped antisense oligonucleotide comprising 2′-O-methyl RNA arms and an unmodified DNA gap was synthesized. A complementary oligonucleotide was also synthesized using unmodified RNA. A double-stranded duplex was formed and the composition was found to inhibit expression of the target gene.
example 2
Length of Double-Stranded Oligonucleotides and the Presence or Absence of Overhangs has No Effect on Function
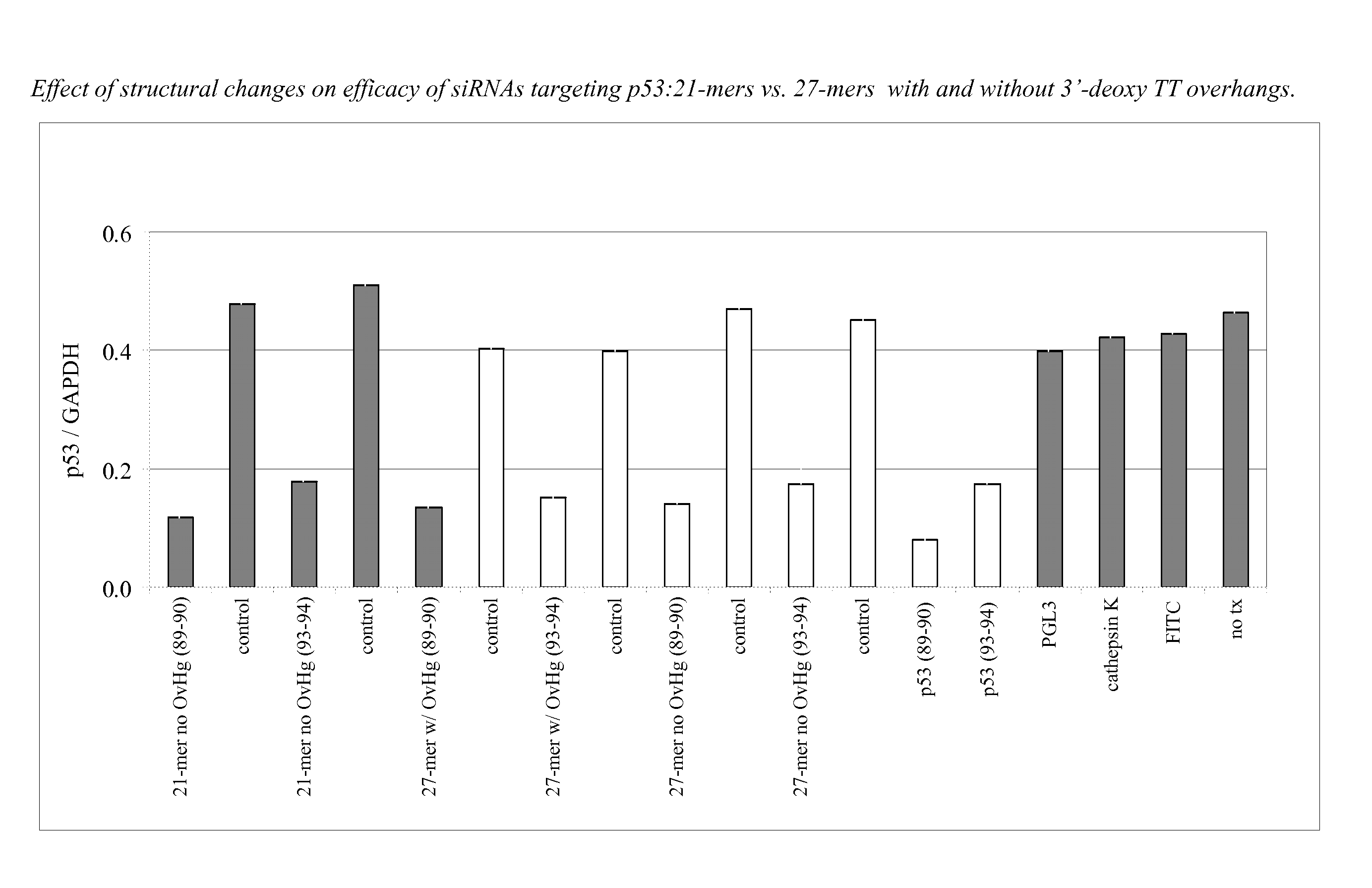
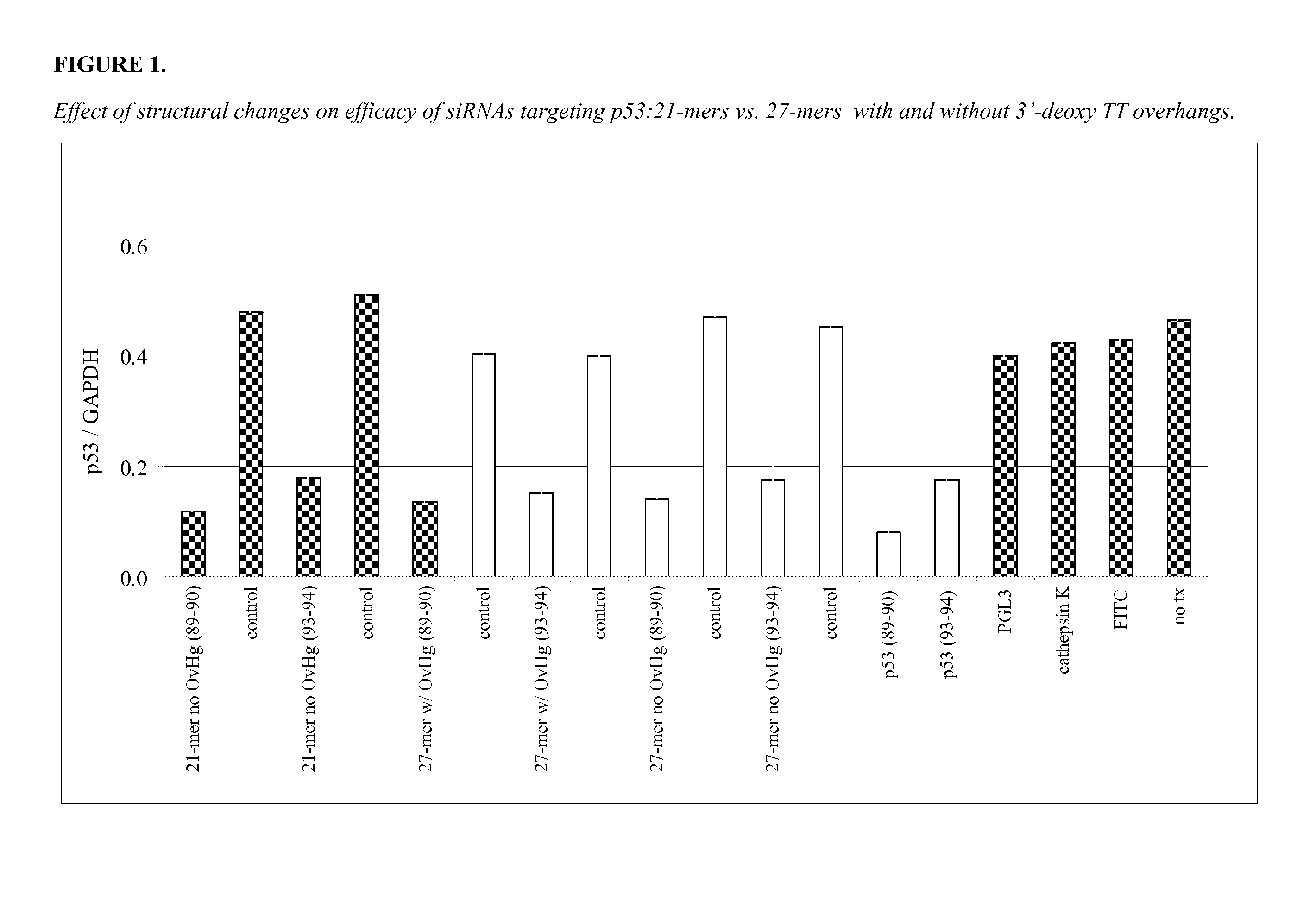
[0353]Twenty one and 27-mers were designed to target each of two sites on the p53 molecule (89-90 site, and 93-94 site). The double-stranded molecules were designed with or without 3′-deoxy TT overhangs. The test oligonucleotides were 21-mers with 2 nucleotide 3′ deoxy TT overhangs and without overhangs (blunt ends); and 27-mers with 2 nucleotide 3′ deoxy TT overhangs and without overhangs (blunt ends). Two positive controls were included in the experiment (p53) and two negative controls were also included (FITC).
[0354]A549 cells were transfected with 100 nM of the double-stranded molecules plus 2 ug / mL LIPOFECTAMINE™ 2000. A549 cells were examined 24 hours post-transfection. FITC-labeled molecules were taken up well by cells. Both 21-mers (with or without overhangs) and 27-mers (with or without overhangs) were non-toxic to cells. FIG. 1 shows the result of an experiment comp...
example 3
Activation of the Double-Stranded RNA, Interferon-Inducible Protein Kinase, PKR
[0356]PKR is activated by double-stranded RNA molecules. Active PKR leads to the inhibition of protein synthesis, activation of transcription, and a variety of other cellular effects, including signal transduction, cell differentiation, cell growth inhibition, apoptosis, and antiviral effects. The effect of p53-targeted double-stranded RNA molecules on PKR expression was tested. The level of mRNA was determined using RT-PCR analysis. As shown in FIG. 2, no correlation was observed between the length of the double-stranded oligonucleotide and the level of PKR induction. Accordingly, long oligonucleotides can be used without activating PKR, a marker for interferon induction.
[0357]As illustrated in FIG. 2B, analysis of relative amounts of PKR mRNA after the 21- and 27-mer transfection in HMVEC cells showed approximately a 2 fold increase in PKR mRNA of the siRNA control sequences over no treatment, and appro...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com