G protein-coupled receptor structural model and a method of designing ligand binding to g protein-coupled receptor by using the structural model
a g protein-coupled receptor and structural model technology, applied in the field of structural models, can solve the problems of rhodopsin not solely providing a structural model, the role of the kink in the functioning of gpcrs remains unclear, and the nature of specific conformational changes has yet to be understood
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of Models for Photoactivated Intermediates of Rhodopsin
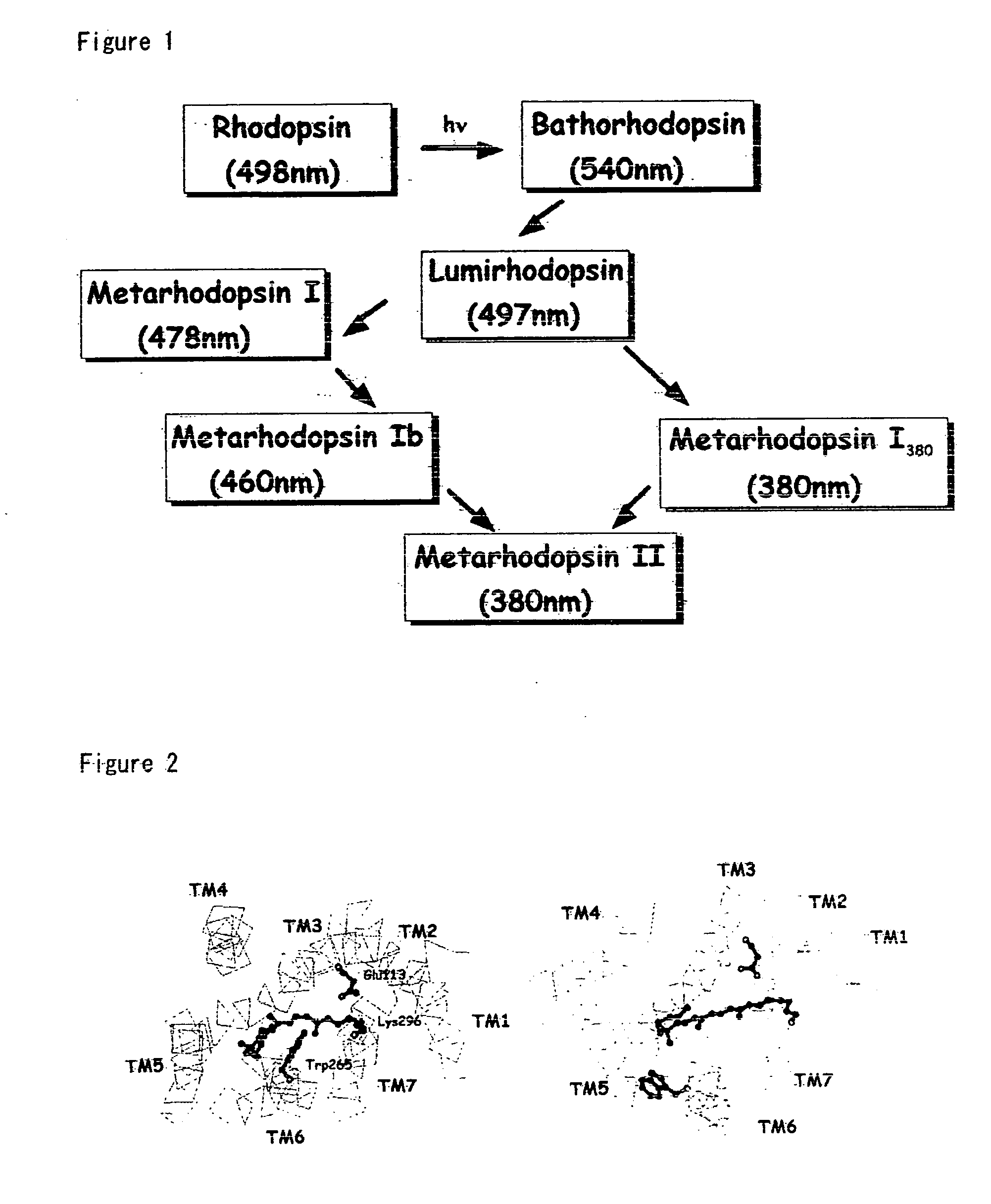
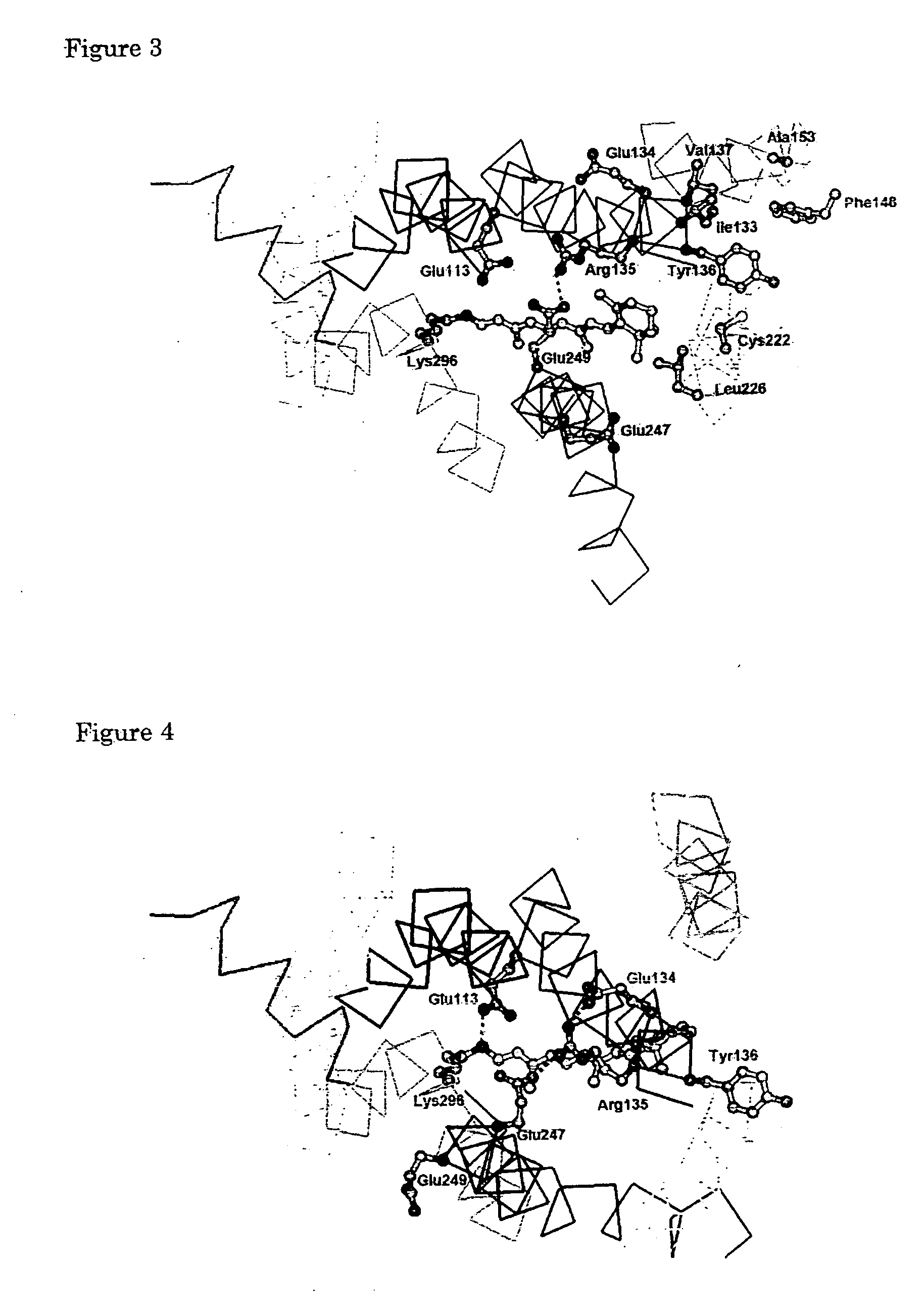
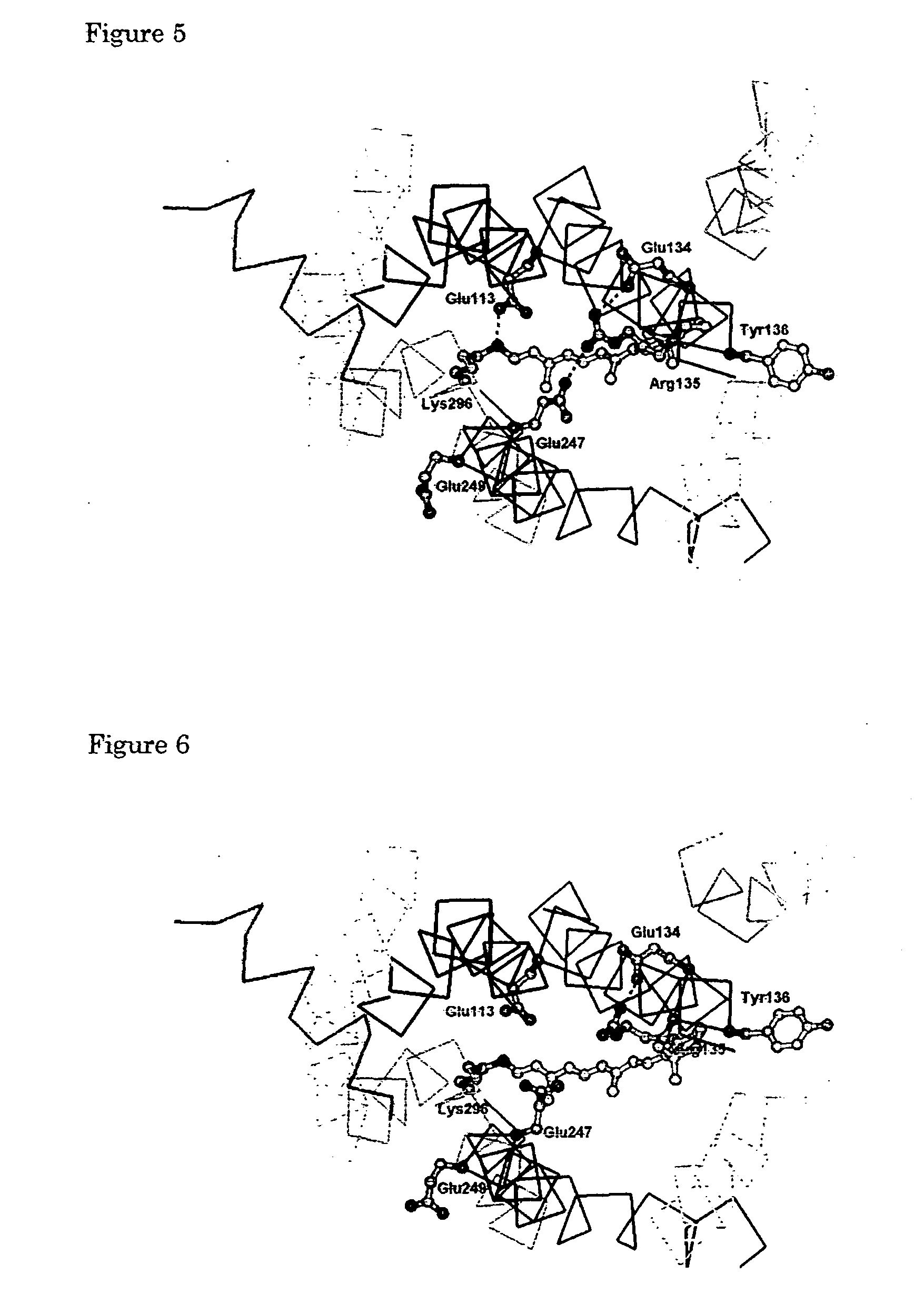
[0126] Using a molecule modeling software Insight II-Discover 3 (Molecular Simulations Inc., USA), a structural model for each of the rhodopsin intermediates was generated and was optimized based an the crystal structure of rhodopsin (Palczewski et al., Science, 289, 144-167, 2000). TM3 was swung about the C α carbon of Cys110 to serve as the pivot point while the distance to TM2 was kept at 5 Å or more. The magnitude of the swing was determined by taking into consideration the interaction of TM6 with Glu247 for each of Lumi, Meta I, Meta Ib, and Meta I380 structures. Specifically, in each of Lumi, Meta I, Meta Ib, and Meta I380, Cys140 on TM3 was swung in such a manner that Cys140 is spaced form TM6 by a distance of 1.6 Å, 4.3 Å, 6.8 Å, and 9.0 Å, respectively. Furthermore, N-terminal (Glu150) of the portion of TM4 that would interfere with TM3 was swung toward TM5 about Gly174 on the C-terminal of the helix to se...
example 2
Construction of models for GPCR and GPCR / Ligand Complex
[0129] Using the structure of Meta I, Meta Ib, Meta I380, and Meta II and based on the homology among the amino acid sequences of rhodopsin and other GPCRs (FIG. 18), three-dimensional conformations for binding a full agonist, a partial agonist, an antagonist, and an inverse agonist were constructed for each of the GPCRs.
[0130] For each of the GPCRs, a receptor conformation for binding an inverse agonist was generated by using the structure of Meta I as a template. Using a homology module of Insight II, amino acid substitution was carried out, as were insertion or deletion of amino acid residues in the loop region. Using Discover 3, the conformation was optimized so that the C α carbon of the amino acids was fixed as firmly as possible.
[0131] Likewise, three-dimensional conformations for binding an antagonist, a partial agonist, and a full agonist that correspond to Meta Ib, Meta I380, and Meta II, respectively, were construc...
example 3
Construction of Structural Model for Adrenaline Receptors Bound to Antagonist
[0133] Using the structure of rhodopsin Meta Ib as a template, Meta Ib-Like structural models of antagonist-bound receptor was constructed for a panel of twelve adrenaline receptors, which form a class of G protein-coupled receptors (GPCRs).
[0134] To construct the structural model for the panel of adrenaline receptors, the amino acid sequence of rhodopsin to serve as a template was first aligned with the amino acid sequences of the panel of adrenaline receptors for which to construct the structural model Clustal W was used as the alignment program (Thompson et al., Nucleic Acids Research, 22:4673-4680(1994)). The analysis revealed that while the amino acid sequences showed a relatively low homology to one another, the transmembrane regions, which include conserved hydrophobic residues and sequence motifs, are aligned at a relatively high homology, and the less conserved loop regions tend to include abnorm...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Crystal structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com