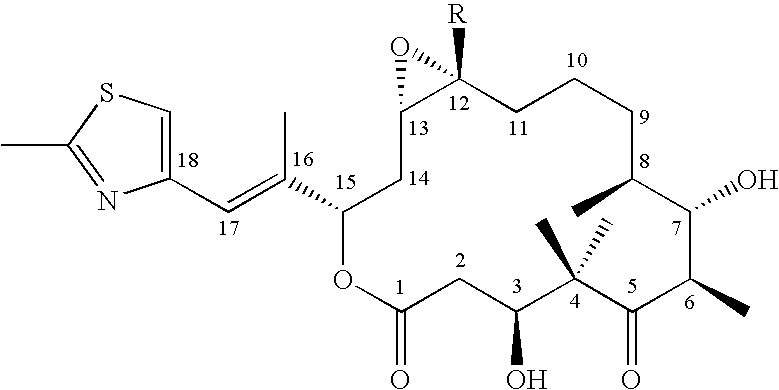
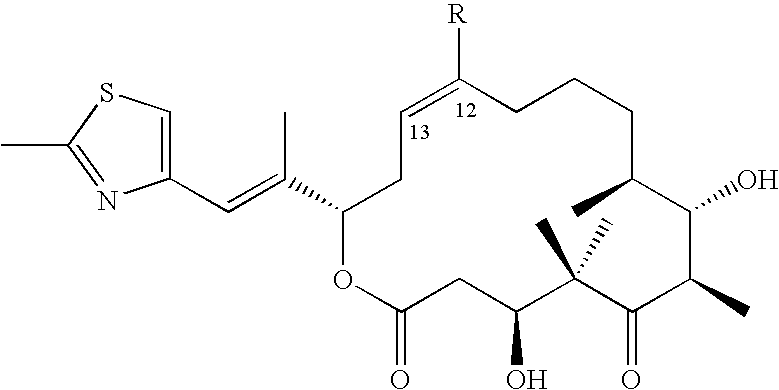
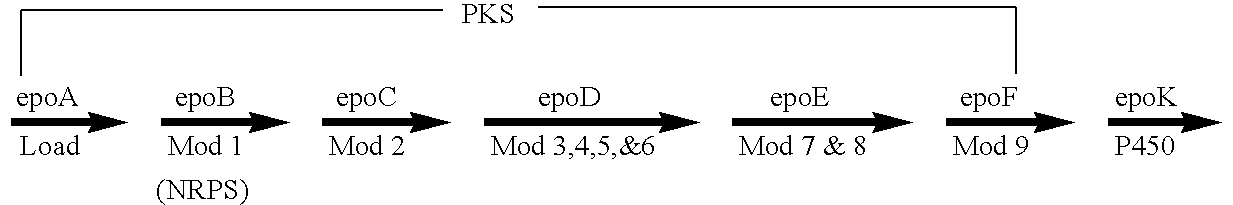
Production of polyketides
a polyketide and recombinant technology, applied in the field of recombinant methods and materials for producing polyketides, can solve the problems of affecting the production of polyketides,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of a Myxococcus xanthus Expression Vector
[0176] The DNA providing the integration and attachment function of phage Mx8 was inserted into commercially available pACYC184 (New England Biolabs). An ˜2360 bp MfeI-SmaI from plasmid pPLH343, described in Salmi et al., February 1998, J. Bact. 180(3): 614-621, was isolated and ligated to the large EcoRI-XmnI restriction fragment of plasmid pACYC184. The circular DNA thus formed was ˜6 kb in size and called plasmid pKOS35-77.
[0177] Plasmid pKOS35-77 serves as a convenient plasmid for expressing recombinant PKS genes of the invention under the control of the epothilone PKS gene promoter. In one illustrative embodiment, the entire epothilone PKS gene with its homologous promoter is inserted in one or more fragments into the plasmid to yield an expression vector of the invention.
[0178] The present invention also provides expression vectors in which the recombinant PKS genes of the invention are under the control of a Myxococcus ...
example 2
Construction of a Bacterial Artificial Chromosome (BAC) for Expression of Epothilone in Myxococcus xanthus
[0185] To express the epothilone PKS and modification enzyme genes in a heterologous host to produce epothilones by fermentation, Myxococcus xanthus, which is closely related to Sorangium cellulosum and for which a number of cloning vectors are available, is employed in accordance with the methods of the invention. M. xanthus and S. cellulosum are myxobacteria and so may share common elements of gene expression, translational control, and post translational modification (if any). M. xanthus has been developed for gene cloning and expression: DNA can be introduced by electroporation, and a number of vectors and genetic markers are available for the introduction of foreign DNA, including those that permit its stable insertion into the chromosome. M. xanthus can be grown with relative ease in complex media in fermentors and can be subjected to manipulations to increase gene expres...
example 3
Process for the Production of Epothilones B and D
A. Production of Epothilone B
[0197] I. Flasks
[0198] A 1 mL vial of the K111-32-25 strain is thawed and the contents transferred into 3 mL of CYE seed media in a glass tube. This culture is incubated for 72±12 hours at 30° C., followed by the subculturing of 3 mL of this tube culture into 50 mL of CYE media within a 250 mL baffled Erlenmeyer flask. This CYE flask is incubated for 24±8 hours at 30° C., and 2.5 mL of this seed (5% v / v) used to inoculate the epothilone production flasks (50 mL of CTS-TA media in a 250 mL baffled Erlenmeyer flask). These flasks are then incubated at 30° C. for 48±12 hours, with a media pH at the beginning of 7.4. The peak epothilone A titer is 0.5 mg / L, and the peak epothilone B titer is 2.5 mg / L.
[0199] II. Fermentors
[0200] A similar inoculum expansion of K111-32-25 as described above is used, with the additional step that 25 mL of the 50 mL CYE seed is subcultured into 500 mL of CYE. This secondary ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| total volume | aaaaa | aaaaa |
| total volume | aaaaa | aaaaa |
| total volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com